Microtubule-based nuclear movement occurs independently of centrosome positioning in migrating neurons
- PMID: 17913873
- PMCID: PMC2000450
- DOI: 10.1073/pnas.0708047104
Microtubule-based nuclear movement occurs independently of centrosome positioning in migrating neurons
Abstract
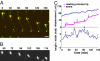
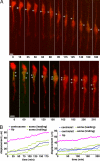
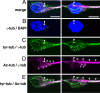
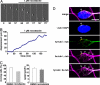
During neuronal migration in the developing brain, it is thought that the centrosome precedes the nucleus and provides a cue for nuclear migration along the microtubules. In time-lapse imaging studies of radially migrating granule cells in mouse cerebellar slices, we observed that the movements of the nucleus and centrosome appeared to occur independently of each other. The nucleus often migrated ahead of the centrosome during its saltatory movement, negating the supposed role of the centrosome in pulling the nucleus. The nucleus was associated with dynamic microtubules enveloping the entire nucleus and stable microtubules extending from the leading process to the anterior part of the nucleus. Neither of these perinuclear microtubules converged at the centrosome. Disruption or excess formation of stable microtubules attenuated nuclear migration, indicating that the configuration of stable microtubules is crucial for nuclear migration. The inhibition of LIS1 function, a regulator of a microtubule motor dynein, specifically blocks nuclear migration without affecting the coupling of the centrosome and microtubules in the leading process, suggesting that movements of the nucleus and centrosome are differentially regulated by dynein motor function. Thus, the nucleus moves along the microtubules independently of the position of the centrosome in migrating neurons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration.J Cell Biol. 2004 Jun 7;165(5):709-21. doi: 10.1083/jcb.200309025. Epub 2004 Jun 1. J Cell Biol. 2004. PMID: 15173193 Free PMC article.
-
Distinct dose-dependent cortical neuronal migration and neurite extension defects in Lis1 and Ndel1 mutant mice.J Neurosci. 2009 Dec 9;29(49):15520-30. doi: 10.1523/JNEUROSCI.4630-09.2009. J Neurosci. 2009. PMID: 20007476 Free PMC article.
-
Nesprins and opposing microtubule motors generate a point force that drives directional nuclear motion in migrating neurons.Development. 2018 Mar 8;145(5):dev158782. doi: 10.1242/dev.158782. Development. 2018. PMID: 29519888
-
Nucleokinesis in neuronal migration.Neuron. 2005 May 5;46(3):383-8. doi: 10.1016/j.neuron.2005.04.013. Neuron. 2005. PMID: 15882636 Review.
-
[Molecular mechanism of lissencephaly--how LIS1 and NDEL1 regulate cytoplasmic dynein?].Brain Nerve. 2008 Apr;60(4):375-81. Brain Nerve. 2008. PMID: 18421979 Review. Japanese.
Cited by
-
Semaphorins deployed to repel cell migrants at spinal cord borders.J Biol. 2008;7(2):4. doi: 10.1186/jbiol65. J Biol. 2008. PMID: 18279538 Free PMC article. Review.
-
Drebrin-mediated microtubule-actomyosin coupling steers cerebellar granule neuron nucleokinesis and migration pathway selection.Nat Commun. 2017 Feb 23;8:14484. doi: 10.1038/ncomms14484. Nat Commun. 2017. PMID: 28230156 Free PMC article.
-
Doublecortin restricts neuronal branching by regulating tubulin polyglutamylation.Nat Commun. 2025 Feb 18;16(1):1749. doi: 10.1038/s41467-025-56951-2. Nat Commun. 2025. PMID: 39966472 Free PMC article.
-
Prickle1b mediates interpretation of migratory cues during zebrafish facial branchiomotor neuron migration.Dev Dyn. 2010 Jun;239(6):1596-608. doi: 10.1002/dvdy.22283. Dev Dyn. 2010. PMID: 20503357 Free PMC article.
-
Astn2, a novel member of the astrotactin gene family, regulates the trafficking of ASTN1 during glial-guided neuronal migration.J Neurosci. 2010 Jun 23;30(25):8529-40. doi: 10.1523/JNEUROSCI.0032-10.2010. J Neurosci. 2010. PMID: 20573900 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

