Protein stability imposes limits on organism complexity and speed of molecular evolution
- PMID: 17913881
- PMCID: PMC2042177
- DOI: 10.1073/pnas.0705366104
Protein stability imposes limits on organism complexity and speed of molecular evolution
Abstract
Classical population genetics a priori assigns fitness to alleles without considering molecular or functional properties of proteins that these alleles encode. Here we study population dynamics in a model where fitness can be inferred from physical properties of proteins under a physiological assumption that loss of stability of any protein encoded by an essential gene confers a lethal phenotype. Accumulation of mutations in organisms containing Gamma genes can then be represented as diffusion within the Gamma-dimensional hypercube with adsorbing boundaries determined, in each dimension, by loss of a protein's stability and, at higher stability, by lack of protein sequences. Solving the diffusion equation whose parameters are derived from the data on point mutations in proteins, we determine a universal distribution of protein stabilities, in agreement with existing data. The theory provides a fundamental relation between mutation rate, maximal genome size, and thermodynamic response of proteins to point mutations. It establishes a universal speed limit on rate of molecular evolution by predicting that populations go extinct (via lethal mutagenesis) when mutation rate exceeds approximately six mutations per essential part of genome per replication for mesophilic organisms and one to two mutations per genome per replication for thermophilic ones. Several RNA viruses function close to the evolutionary speed limit, whereas error correction mechanisms used by DNA viruses and nonmutant strains of bacteria featuring various genome lengths and mutation rates have brought these organisms universally approximately 1,000-fold below the natural speed limit.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
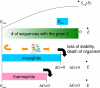
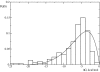

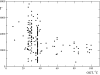
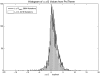
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

