Organization of sister origins and replisomes during multifork DNA replication in Escherichia coli
- PMID: 17914458
- PMCID: PMC2063475
- DOI: 10.1038/sj.emboj.7601871
Organization of sister origins and replisomes during multifork DNA replication in Escherichia coli
Abstract
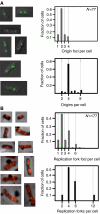
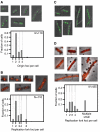
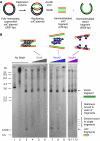
The replication period of Escherichia coli cells grown in rich medium lasts longer than one generation. Initiation thus occurs in the 'mother-' or 'grandmother generation'. Sister origins in such cells were found to be colocalized for an entire generation or more, whereas sister origins in slow-growing cells were colocalized for about 0.1-0.2 generations. The role of origin inactivation (sequestration) by the SeqA protein in origin colocalization was studied by comparing sequestration-deficient mutants with wild-type cells. Cells with mutant, non-sequesterable origins showed wild-type colocalization of sister origins. In contrast, cells unable to sequester new origins due to loss of SeqA, showed aberrant localization of origins indicating a lack of organization of new origins. In these cells, aberrant replisome organization was also found. These results suggest that correct organization of sister origins and sister replisomes is dependent on the binding of SeqA protein to newly formed DNA at the replication forks, but independent of origin sequestration. In agreement, in vitro experiments indicate that SeqA is capable of pairing newly replicated DNA molecules.
Figures
Similar articles
-
Escherichia coli SeqA structures relocalize abruptly upon termination of origin sequestration during multifork DNA replication.PLoS One. 2014 Oct 21;9(10):e110575. doi: 10.1371/journal.pone.0110575. eCollection 2014. PLoS One. 2014. PMID: 25333813 Free PMC article.
-
Dynamic Escherichia coli SeqA complexes organize the newly replicated DNA at a considerable distance from the replisome.Nucleic Acids Res. 2015 Mar 11;43(5):2730-43. doi: 10.1093/nar/gkv146. Epub 2015 Feb 26. Nucleic Acids Res. 2015. PMID: 25722374 Free PMC article.
-
Lack of SeqA focus formation, specific DNA binding and proper protein multimerization in the Escherichia coli sequestration mutant seqA2.Mol Microbiol. 2003 Feb;47(3):619-32. doi: 10.1046/j.1365-2958.2003.t01-1-03329.x. Mol Microbiol. 2003. PMID: 12535065
-
The Escherichia coli SeqA protein.Plasmid. 2009 May;61(3):141-50. doi: 10.1016/j.plasmid.2009.02.004. Epub 2009 Feb 28. Plasmid. 2009. PMID: 19254745 Review.
-
Partitioning of the Escherichia coli chromosome: superhelicity and condensation.Biochimie. 2001 Jan;83(1):41-8. doi: 10.1016/s0300-9084(00)01204-9. Biochimie. 2001. PMID: 11254973 Review.
Cited by
-
Knockdown Experiment Reveals an Essential GTPase CgtA's Involvement in Growth, Viability, Motility, Morphology, and Persister Phenotypes in Vibrio cholerae.Microbiol Spectr. 2023 Mar 14;11(2):e0318122. doi: 10.1128/spectrum.03181-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36916969 Free PMC article.
-
DNA motifs that sculpt the bacterial chromosome.Nat Rev Microbiol. 2011 Jan;9(1):15-26. doi: 10.1038/nrmicro2477. Nat Rev Microbiol. 2011. PMID: 21164534 Review.
-
Protein aggregation in E. coli : short term and long term effects of nutrient density.PLoS One. 2014 Sep 11;9(9):e107445. doi: 10.1371/journal.pone.0107445. eCollection 2014. PLoS One. 2014. PMID: 25210787 Free PMC article.
-
Rhodoccoccus erythropolis Is Different from Other Members of Actinobacteria: Monoploidy, Overlapping Replication Cycle, and Unique Segregation Pattern.J Bacteriol. 2019 Nov 20;201(24):e00320-19. doi: 10.1128/JB.00320-19. Print 2019 Dec 15. J Bacteriol. 2019. PMID: 31570531 Free PMC article.
-
Multifork chromosome replication in slow-growing bacteria.Sci Rep. 2017 Mar 6;7:43836. doi: 10.1038/srep43836. Sci Rep. 2017. PMID: 28262767 Free PMC article.
References
-
- Bach T, Skarstad K (2004) Re-replication from non-sequesterable origins generates three-nucleoid cells which divide asymmetrically. Mol Microbiol 51: 1589–1600 - PubMed
-
- Berkmen MB, Grossman AD (2006) Spatial and temporal organization of the Bacillus subtilis replication cycle. Mol Microbiol 62: 57–71 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

