Measurement of the incorporation and repair of exogenous 5-hydroxymethyl-2'-deoxyuridine in human cells in culture using gas chromatography-negative chemical ionization-mass spectrometry
- PMID: 17914883
- PMCID: PMC2532841
- DOI: 10.1021/tx700221x
Measurement of the incorporation and repair of exogenous 5-hydroxymethyl-2'-deoxyuridine in human cells in culture using gas chromatography-negative chemical ionization-mass spectrometry
Abstract
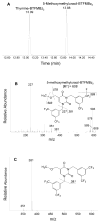
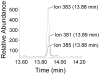
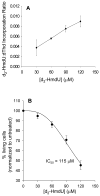
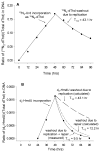
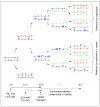
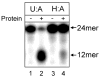
The DNA of all organisms is constantly damaged by oxidation. Among the array of damage products is 5-hydroxymethyluracil, derived from oxidation of the thymine methyl group. Previous studies have established that HmU can be a sensitive and valuable marker of DNA damage. More recently, the corresponding deoxynucleoside, 5-hydroxymethyl-2'-deoxyuridine (HmdU), has proven to be valuable for the introduction of controlled amounts of a single type of damage lesion into the DNA of replicating cells, which is subsequently repaired by the base excision repair pathway. Complicating the study of HmU formation and repair, however, is the known chemical reactivity of the hydroxymethyl group of HmU under conditions used to hydrolyze DNA. In the work reported here, this chemical property has been exploited by creating conditions that convert HmU to the corresponding methoxymethyluracil (MmU) derivative that can be further derivatized to the 3,5-bis-(trifluoromethyl)benzyl analogue. This derivatized compound can be detected by gas chromatography-negative chemical ionization-mass spectrometry (GC-NCI-MS) with good sensitivity. Using isotopically enriched exogenous HmdU and human osteosarcoma cells (U2OS) in culture, we demonstrate that this method allows for the measurement of HmU in DNA formed from the incorporation of exogenous HmdU. We further demonstrate that the addition of isotopically enriched uridine to the culture medium allows for the simultaneous measurement of DNA replication and repair kinetics. This sensitive and facile method should prove valuable for studies on DNA oxidation damage and repair in living cells.
Figures
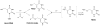
Similar articles
-
Quantification of 5-(hydroxymethyl)uracil in DNA by gas chromatography/mass spectrometry: problems and solutions.Chem Res Toxicol. 1998 Jul;11(7):786-93. doi: 10.1021/tx970233c. Chem Res Toxicol. 1998. PMID: 9671541
-
Measurement of urinary excretion of 5-hydroxymethyluracil in human by GC/NICI/MS: correlation with cigarette smoking, urinary TBARS and etheno DNA adduct.Toxicol Lett. 2005 Mar 15;155(3):403-10. doi: 10.1016/j.toxlet.2004.11.009. Toxicol Lett. 2005. PMID: 15649624
-
Observation and prevention of an artefactual formation of oxidized DNA bases and nucleosides in the GC-EIMS method.Carcinogenesis. 1996 Feb;17(2):347-53. doi: 10.1093/carcin/17.2.347. Carcinogenesis. 1996. PMID: 8625462
-
Facts about the artifacts in the measurement of oxidative DNA base damage by gas chromatography-mass spectrometry.Free Radic Res. 1998 Dec;29(6):551-63. doi: 10.1080/10715769800300591. Free Radic Res. 1998. PMID: 10098459 Review.
-
Substrate specificities and excision kinetics of DNA glycosylases involved in base-excision repair of oxidative DNA damage.Mutat Res. 2003 Oct 29;531(1-2):109-26. doi: 10.1016/j.mrfmmm.2003.07.003. Mutat Res. 2003. PMID: 14637249 Review.
Cited by
-
The effect of Pot1 binding on the repair of thymine analogs in a telomeric DNA sequence.Nucleic Acids Res. 2014 Aug;42(14):9063-73. doi: 10.1093/nar/gku602. Epub 2014 Jul 22. Nucleic Acids Res. 2014. PMID: 25053838 Free PMC article.
-
Mass spectrometry of structurally modified DNA.Chem Rev. 2013 Apr 10;113(4):2395-436. doi: 10.1021/cr300391r. Epub 2013 Feb 26. Chem Rev. 2013. PMID: 23441727 Free PMC article. Review. No abstract available.
-
DNA Base Excision Repair Intermediates Influence Duplex-Quadruplex Equilibrium.Molecules. 2023 Jan 18;28(3):970. doi: 10.3390/molecules28030970. Molecules. 2023. PMID: 36770637 Free PMC article.
-
Synergistic enhancement of 5-fluorouracil cytotoxicity by deoxyuridine analogs in cancer cells.Oncoscience. 2015 Feb 9;2(3):272-84. doi: 10.18632/oncoscience.125. eCollection 2015. Oncoscience. 2015. PMID: 25897430 Free PMC article.
References
-
- Loeb LA. Endogenous carcinogenesis: molecular oncology into the twenty-first century – presidential address. Cancer Res. 1989;49:5489–5496. - PubMed
-
- Mullaart E, Lohman PH, Berends F, Vijg J. DNA damage metabolism and aging. Mutat Res. 1990;237:189–210. - PubMed
-
- Frenkel K, Zhong ZJ, Wei HC, Karkoszka J, Patel U, Rashid K, Georgescu M, Solomon JJ. Quantitative high-performance liquid chromatography analysis of DNA oxidized in vitro and in vivo. Anal Biochem. 1991;196:126–136. - PubMed
-
- Djuric Z, Luongo DA, Harper DA. Quantitation of 5-(hydroxymethyl)uracil in DNA by gas chromatography with mass spectral detection. Chem Res Toxicol. 1991;4:687–691. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

