Solution structure of polymerase mu's BRCT Domain reveals an element essential for its role in nonhomologous end joining
- PMID: 17915942
- PMCID: PMC2653216
- DOI: 10.1021/bi7007728
Solution structure of polymerase mu's BRCT Domain reveals an element essential for its role in nonhomologous end joining
Abstract
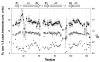
The solution structure and dynamics of the BRCT domain from human DNA polymerase mu, implicated in repair of chromosome breaks by nonhomologous end joining (NHEJ), has been determined using NMR methods. BRCT domains are typically involved in protein-protein interactions between factors required for the cellular response to DNA damage. The pol mu BRCT domain is atypical in that, unlike other reported BRCT structures, the pol mu BRCT is neither part of a tandem grouping, nor does it appear to form stable homodimers. Although the sequence of the pol mu BRCT domain has some unique characteristics, particularly the presence of >10% proline residues, it forms the characteristic alphabetaalpha sandwich, in which three alpha helices are arrayed around a central four-stranded beta-sheet. The structure of helix alpha1 is characterized by two solvent-exposed hydrophobic residues, F46 and L50, suggesting that this element may play a role in mediating interactions of pol mu with other proteins. Consistent with this argument, mutation of these residues, as well as the proximal, conserved residue R43, specifically blocked the ability of pol mu to efficiently work together with NHEJ factors Ku and XRCC4-ligase IV to join noncomplementary ends together in vitro. The structural, dynamic, and biochemical evidence reported here identifies a functional surface in the pol mu BRCT domain critical for promoting assembly and activity of the NHEJ machinery. Further, the similarity between the interaction regions of the BRCT domains of pol mu and TdT support the conclusion that they participate in NHEJ as alternate polymerases.
Figures

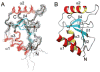
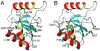



References
-
- Fan W, Wu X. DNA polymerase lambda can elongate on DNA substrates mimicking non-homologous end joining and interact with XRCC4-ligase IV complex. Biochem Biophys Res Commun. 2004;323:1328–1333. - PubMed
-
- Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase λ in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J Biol Chem. 2004;279:805–811. - PubMed
-
- Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell. 2004;16:701–713. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

