All duplicates are not equal: the difference between small-scale and genome duplication
- PMID: 17916239
- PMCID: PMC2246283
- DOI: 10.1186/gb-2007-8-10-r209
All duplicates are not equal: the difference between small-scale and genome duplication
Abstract
Background: Genes in populations are in constant flux, being gained through duplication and occasionally retained or, more frequently, lost from the genome. In this study we compare pairs of identifiable gene duplicates generated by small-scale (predominantly single-gene) duplications with those created by a large-scale gene duplication event (whole-genome duplication) in the yeast Saccharomyces cerevisiae.
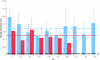
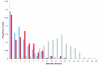
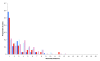
Results: We find a number of quantifiable differences between these data sets. Whole-genome duplicates tend to exhibit less profound phenotypic effects when deleted, are functionally less divergent, and are associated with a different set of functions than their small-scale duplicate counterparts. At first sight, either of these latter two features could provide a plausible mechanism by which the difference in dispensability might arise. However, we uncover no evidence suggesting that this is the case. We find that the difference in dispensability observed between the two duplicate types is limited to gene products found within protein complexes, and probably results from differences in the relative strength of the evolutionary pressures present following each type of duplication event.
Conclusion: Genes, and the proteins they specify, originating from small-scale and whole-genome duplication events differ in quantifiable ways. We infer that this is not due to their association with different functional categories; rather, it is a direct result of biases in gene retention.
Figures
Similar articles
-
Variation in gene duplicates with low synonymous divergence in Saccharomyces cerevisiae relative to Caenorhabditis elegans.Genome Biol. 2009;10(7):R75. doi: 10.1186/gb-2009-10-7-r75. Epub 2009 Jul 13. Genome Biol. 2009. PMID: 19594930 Free PMC article.
-
Retention of protein complex membership by ancient duplicated gene products in budding yeast.Trends Genet. 2007 Jun;23(6):266-9. doi: 10.1016/j.tig.2007.03.012. Epub 2007 Apr 10. Trends Genet. 2007. PMID: 17428571 Review.
-
The roles of whole-genome and small-scale duplications in the functional specialization of Saccharomyces cerevisiae genes.PLoS Genet. 2013;9(1):e1003176. doi: 10.1371/journal.pgen.1003176. Epub 2013 Jan 3. PLoS Genet. 2013. PMID: 23300483 Free PMC article.
-
Gene duplication and environmental adaptation within yeast populations.Genome Biol Evol. 2010;2:591-601. doi: 10.1093/gbe/evq043. Epub 2010 Jul 21. Genome Biol Evol. 2010. PMID: 20660110 Free PMC article.
-
Gene duplication as a major force in evolution.J Genet. 2013 Apr;92(1):155-61. doi: 10.1007/s12041-013-0212-8. J Genet. 2013. PMID: 23640422 Review.
Cited by
-
Evolutionary conservation of sequence motifs at sites of protein modification.J Biol Chem. 2023 May;299(5):104617. doi: 10.1016/j.jbc.2023.104617. Epub 2023 Mar 16. J Biol Chem. 2023. PMID: 36933807 Free PMC article.
-
The evolution of vertebrate tetraspanins: gene loss, retention, and massive positive selection after whole genome duplications.BMC Evol Biol. 2010 Oct 13;10:306. doi: 10.1186/1471-2148-10-306. BMC Evol Biol. 2010. PMID: 20939927 Free PMC article.
-
Parallel Concerted Evolution of Ribosomal Protein Genes in Fungi and Its Adaptive Significance.Mol Biol Evol. 2020 Feb 1;37(2):455-468. doi: 10.1093/molbev/msz229. Mol Biol Evol. 2020. PMID: 31589316 Free PMC article.
-
The multiple fates of gene duplications: Deletion, hypofunctionalization, subfunctionalization, neofunctionalization, dosage balance constraints, and neutral variation.Plant Cell. 2022 Jul 4;34(7):2466-2474. doi: 10.1093/plcell/koac076. Plant Cell. 2022. PMID: 35253876 Free PMC article.
-
A Single, Shared Triploidy in Three Species of Parasitic Nematodes.G3 (Bethesda). 2020 Jan 7;10(1):225-233. doi: 10.1534/g3.119.400650. G3 (Bethesda). 2020. PMID: 31694855 Free PMC article.
References
-
- Ohno S. Evolution by Gene Duplication. London, New York: Allen & Unwin, Springer-Verlag; 1970.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

