Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases
- PMID: 17916244
- PMCID: PMC2082048
- DOI: 10.1186/1471-2105-8-372
Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases
Abstract
Background: In recent years, mammalian protein-protein interaction network databases have been developed. The interactions in these databases are either extracted manually from low-throughput experimental biomedical research literature, extracted automatically from literature using techniques such as natural language processing (NLP), generated experimentally using high-throughput methods such as yeast-2-hybrid screens, or interactions are predicted using an assortment of computational approaches. Genes or proteins identified as significantly changing in proteomic experiments, or identified as susceptibility disease genes in genomic studies, can be placed in the context of protein interaction networks in order to assign these genes and proteins to pathways and protein complexes.
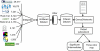
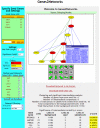
Results: Genes2Networks is a software system that integrates the content of ten mammalian interaction network datasets. Filtering techniques to prune low-confidence interactions were implemented. Genes2Networks is delivered as a web-based service using AJAX. The system can be used to extract relevant subnetworks created from "seed" lists of human Entrez gene symbols. The output includes a dynamic linkable three color web-based network map, with a statistical analysis report that identifies significant intermediate nodes used to connect the seed list.
Conclusion: Genes2Networks is powerful web-based software that can help experimental biologists to interpret lists of genes and proteins such as those commonly produced through genomic and proteomic experiments, as well as lists of genes and proteins associated with disease processes. This system can be used to find relationships between genes and proteins from seed lists, and predict additional genes or proteins that may play key roles in common pathways or protein complexes.
Figures
References
-
- Fields S, Song O-k. A novel genetic system to detect proteinÂ-protein interactions. 1989;340:245–246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

