Regulation of low affinity neurotrophin receptor (p75(NTR)) by early growth response (Egr) transcriptional regulators
- PMID: 17916431
- PMCID: PMC2098703
- DOI: 10.1016/j.mcn.2007.08.013
Regulation of low affinity neurotrophin receptor (p75(NTR)) by early growth response (Egr) transcriptional regulators
Abstract
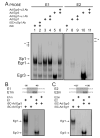
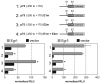
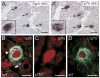
The low affinity neurotrophin receptor p75(NTR) is a multifunctional receptor with important roles in neurotrophin signaling, axon outgrowth, and oligodendroglia and neuron survival. It is transcriptionally regulated with spatial and temporal precision during nervous system development, injury and regeneration. Very little is known about how p75(NTR) expression is dynamically regulated but it is likely to influence how p75(NTR) signals in particular cellular contexts. Here, we identify the early growth response (Egr) transcriptional regulators, Egr1 and Egr3, as direct modulators of p75(NTR) gene expression. Egr1 and Egr3 bind and transactivate the p75(NTR) promoter in vitro and in vivo, using distinct response elements on the p75(NTR) promoter. Consistent with these results, p75(NTR) expression is greatly diminished in muscle spindle stretch receptors and in peripheral nerve Schwann cells in Egr gene deficient mice. Taken together, the results elucidate a novel mechanism whereby Egr proteins can directly modulate p75(NTR) expression and signaling in vivo.
Figures





Similar articles
-
Schwann cell p75NTR prevents spontaneous sensory reinnervation of the adult spinal cord.Brain. 2010 Feb;133(Pt 2):421-32. doi: 10.1093/brain/awp316. Epub 2010 Jan 3. Brain. 2010. PMID: 20047901
-
Merlin status regulates p75(NTR) expression and apoptotic signaling in Schwann cells following nerve injury.Neurobiol Dis. 2015 Oct;82:114-122. doi: 10.1016/j.nbd.2015.05.021. Epub 2015 Jun 6. Neurobiol Dis. 2015. PMID: 26057084 Free PMC article.
-
p75 neurotrophin receptor expression defines a population of BDNF-responsive neurogenic precursor cells.J Neurosci. 2007 May 9;27(19):5146-55. doi: 10.1523/JNEUROSCI.0654-07.2007. J Neurosci. 2007. PMID: 17494700 Free PMC article.
-
p75 neurotrophin receptor signaling in the nervous system.Biotechnol Annu Rev. 2004;10:123-49. doi: 10.1016/S1387-2656(04)10005-7. Biotechnol Annu Rev. 2004. PMID: 15504705 Review.
-
Proteolytic processing of the p75 neurotrophin receptor: A prerequisite for signalling?: Neuronal life, growth and death signalling are crucially regulated by intra-membrane proteolysis and trafficking of p75(NTR).Bioessays. 2011 Aug;33(8):614-25. doi: 10.1002/bies.201100036. Epub 2011 Jun 30. Bioessays. 2011. PMID: 21717487 Review.
Cited by
-
Early growth response 3 (Egr-3) is induced by transforming growth factor-β and regulates fibrogenic responses.Am J Pathol. 2013 Oct;183(4):1197-1208. doi: 10.1016/j.ajpath.2013.06.016. Epub 2013 Jul 30. Am J Pathol. 2013. PMID: 23906810 Free PMC article.
-
P75 neurotrophin receptor controls subventricular zone neural stem cell migration after stroke.Cell Tissue Res. 2022 Mar;387(3):415-431. doi: 10.1007/s00441-021-03539-z. Epub 2021 Oct 26. Cell Tissue Res. 2022. PMID: 34698916 Free PMC article. Review.
-
A novel microRNA and transcription factor mediated regulatory network in schizophrenia.BMC Syst Biol. 2010 Feb 15;4:10. doi: 10.1186/1752-0509-4-10. BMC Syst Biol. 2010. PMID: 20156358 Free PMC article.
-
Molecular Mechanisms Involved in Schwann Cell Plasticity.Front Mol Neurosci. 2017 Feb 17;10:38. doi: 10.3389/fnmol.2017.00038. eCollection 2017. Front Mol Neurosci. 2017. PMID: 28261057 Free PMC article. Review.
-
The molecular machinery of myelin gene transcription in Schwann cells.Glia. 2008 Nov 1;56(14):1541-1551. doi: 10.1002/glia.20767. Glia. 2008. PMID: 18803322 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

