Molecular cloning and characterization of estrogen, androgen, and progesterone nuclear receptors from a freshwater turtle (Pseudemys nelsoni)
- PMID: 17916628
- PMCID: PMC2734501
- DOI: 10.1210/en.2007-0938
Molecular cloning and characterization of estrogen, androgen, and progesterone nuclear receptors from a freshwater turtle (Pseudemys nelsoni)
Abstract
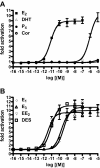
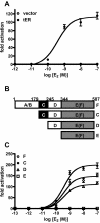
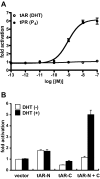
Steroid hormones are essential for the normal function of many organ systems in vertebrates. Reproductive activity in females and males, such as the differentiation, growth, and maintenance of the reproductive system, requires signaling by the sex steroids. Although extensively studied in mammals and a few fish, amphibians, and bird species, the molecular mechanisms of sex steroid hormone (estrogens, androgens, and progestins) action are poorly understood in reptiles. Here we evaluate hormone receptor ligand interactions in a freshwater turtle, the red-belly slider (Pseudemys nelsoni), after the isolation of cDNAs encoding an estrogen receptor alpha (ERalpha), an androgen receptor (AR), and a progesterone receptor (PR). The full-length red-belly slider turtle (t)ERalpha, tAR, and tPR cDNAs were obtained using 5' and 3' rapid amplification cDNA ends. The deduced amino acid sequences showed high identity to the chicken orthologs (tERalpha, 90%; tAR, 71%; tPR, 71%). Using transient transfection assays of mammalian cells, tERalpha protein displayed estrogen-dependent activation of transcription from an estrogen-responsive element-containing promoter. The other receptor proteins, tAR and tPR, also displayed androgen- or progestin-dependent activation of transcription from androgen- and progestin-responsive murine mammary tumor virus promoters. We further examined the transactivation of tERalpha, tAR and tPR by ligands using a modified GAL4-transactivation system. We found that the GAL4-transactivation system was not suitable for the measurement of tAR and tPR transactivations. This is the first report of the full coding regions of a reptilian AR and PR and the examination of their transactivation by steroid hormones.
Figures






References
-
- Blumberg B, Evans RM 1998 Orphan nuclear receptors: new ligands and new possibilities. Gene Dev 12:3149–3155 - PubMed
-
- Katsu Y, Bermudez DS, Braun E, Helbing C, Miyagawa S, Gunderson MP, Kohno S, Bryan TA, Guillette LJ, Iguchi T 2004 Molecular cloning of the estrogen and progesterone receptors of the American alligator. Gen Comp Endocrinol 136:122–133 - PubMed
-
- Katsu Y, Kohno S, Oka T, Mitsui N, Tooi O, Santo N, Urushitani H, Fukumoto Y, Kuwabara K, Ashikaga K, Minami S, Kato S, Ohta Y, Guillette LJ, Iguchi T 2006 Molecular cloning of estrogen receptor α (ERα; ESR1) of the Japanese giant salamander, Andrias japonicus. Mol Cell Endocrinol 257–258:84–94 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

