Breed relationships facilitate fine-mapping studies: a 7.8-kb deletion cosegregates with Collie eye anomaly across multiple dog breeds
- PMID: 17916641
- PMCID: PMC2045139
- DOI: 10.1101/gr.6772807
Breed relationships facilitate fine-mapping studies: a 7.8-kb deletion cosegregates with Collie eye anomaly across multiple dog breeds
Abstract
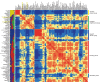
The features of modern dog breeds that increase the ease of mapping common diseases, such as reduced heterogeneity and extensive linkage disequilibrium, may also increase the difficulty associated with fine mapping and identifying causative mutations. One way to address this problem is by combining data from multiple breeds segregating the same trait after initial linkage has been determined. The multibreed approach increases the number of potentially informative recombination events and reduces the size of the critical haplotype by taking advantage of shortened linkage disequilibrium distances found across breeds. In order to identify breeds that likely share a trait inherited from the same ancestral source, we have used cluster analysis to divide 132 breeds of dog into five primary breed groups. We then use the multibreed approach to fine-map Collie eye anomaly (cea), a complex disorder of ocular development that was initially mapped to a 3.9-cM region on canine chromosome 37. Combined genotypes from affected individuals from four breeds of a single breed group significantly narrowed the candidate gene region to a 103-kb interval spanning only four genes. Sequence analysis revealed that all affected dogs share a homozygous deletion of 7.8 kb in the NHEJ1 gene. This intronic deletion spans a highly conserved binding domain to which several developmentally important proteins bind. This work both establishes that the primary cea mutation arose as a single disease allele in a common ancestor of herding breeds as well as highlights the value of comparative population analysis for refining regions of linkage.
Figures


References
-
- Alur R., Brooks B., Brooks B. Clinical and genetic analysis of coloboma: A review. Asian J. Exp. Sci. 2004;20:1–15.
-
- American College of Veterinary Ophthalmologists . Ocular disorders proven or suspected to be inherited in purebred dogs. Genetics Committee Report; Meridian, ID: 2007.
-
- Berryere T.G., Kerns J.A., Barsh G.S., Schmutz S.M., Kerns J.A., Barsh G.S., Schmutz S.M., Barsh G.S., Schmutz S.M., Schmutz S.M. Association of an Agouti allele with fawn or sable coat color in domestic dogs. Mamm. Genome. 2005;16:262–272. - PubMed
-
- Chandler K. Canine epilepsy: What can we learn from human seizure disorders? Vet. J. 2006;172:207–217. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
