Stress-induced mutagenesis in bacteria
- PMID: 17917873
- PMCID: PMC2747772
- DOI: 10.1080/10409230701648494
Stress-induced mutagenesis in bacteria
Abstract
Bacteria spend their lives buffeted by changing environmental conditions. To adapt to and survive these stresses, bacteria have global response systems that result in sweeping changes in gene expression and cellular metabolism. These responses are controlled by master regulators, which include: alternative sigma factors, such as RpoS and RpoH; small molecule effectors, such as ppGpp; gene repressors such as LexA; and, inorganic molecules, such as polyphosphate. The response pathways extensively overlap and are induced to various extents by the same environmental stresses. These stresses include nutritional deprivation, DNA damage, temperature shift, and exposure to antibiotics. All of these global stress responses include functions that can increase genetic variability. In particular, up-regulation and activation of error-prone DNA polymerases, down-regulation of error-correcting enzymes, and movement of mobile genetic elements are common features of several stress responses. The result is that under a variety of stressful conditions, bacteria are induced for genetic change. This transient mutator state may be important for adaptive evolution.
Figures
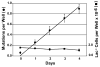
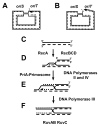

References
-
- Aertsen A, Michiels CW. Mrr instigates the SOS response after high pressure stress in Escherichia coli. Molec Microbiol. 2005;58:1381–1391. - PubMed
-
- Aertsen A, Michiels CW. Upstream of the SOS response: figure out the trigger. Trends Microbiol. 2006;14:421–423. - PubMed
-
- Aguirre-Ramirez M, Ramirez-Santos J, Van ML, Gomez-Eichelmann MC. Expression of the F plasmid ccd toxin-antitoxin system in Escherichia coli cells under nutritional stress. Can J Microbiol. 2006;52:24–30. - PubMed
-
- Al Mamun AA, Marians KJ, Humayun MZ. DNA polymerase III from Escherichia coli cells expressing mutA mistranslator tRNA Is error-prone. J Biol Chem. 2002;277:46319–46327. - PubMed
-
- Andersson DI, Slechta ES, Roth JR. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science. 1998;282:1133–1135. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
