An artificial beta-sheet that dimerizes through parallel beta-sheet interactions
- PMID: 17918935
- PMCID: PMC2593858
- DOI: 10.1021/ja073391r
An artificial beta-sheet that dimerizes through parallel beta-sheet interactions
Abstract
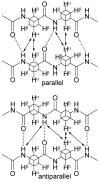
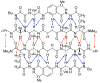
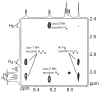
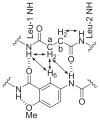
This Article introduces a simple chemical model of a beta-sheet (artificial beta-sheet) that dimerizes by parallel beta-sheet formation in chloroform solution. The artificial beta-sheet consists of two N-terminally linked peptide strands that are linked with succinic or fumaric acid and blocked along one edge with a hydrogen-bonding template composed of 5-aminoanisic acid hydrazide. The template is connected to one of the peptide strands by a turn unit composed of (S)-2-aminoadipic acid (Aaa). 1H NMR spectroscopic studies show that these artificial beta-sheets fold in CDCl3 solution to form well-defined beta-sheet structures that dimerize through parallel beta-sheet interactions. Most notably, all of these compounds show a rich network of NOEs associated with folding and dimerization. The compounds also exhibit chemical shifts and coupling constants consistent with the formation of folded dimeric beta-sheet structures. The aminoadipic acid unit shows patterns of NOEs and coupling constants consistent with a well-defined turn conformation. The present system represents a significant step toward modeling the type of parallel beta-sheet interactions that occur in protein aggregation.
Figures





Similar articles
-
Exploring beta-sheet structure and interactions with chemical model systems.Acc Chem Res. 2008 Oct;41(10):1319-30. doi: 10.1021/ar800064f. Epub 2008 Sep 18. Acc Chem Res. 2008. PMID: 18798654 Free PMC article.
-
A new artificial beta-sheet that dimerizes through parallel beta-sheet interactions.Org Lett. 2009 Feb 19;11(4):1003-6. doi: 10.1021/ol802993v. Org Lett. 2009. PMID: 19173616 Free PMC article.
-
An unnatural amino acid that induces beta-sheet folding and interaction in peptides.J Am Chem Soc. 2002 May 8;124(18):4972-3. doi: 10.1021/ja025699i. J Am Chem Soc. 2002. PMID: 11982357
-
Molecular structures of amyloid and prion fibrils: consensus versus controversy.Acc Chem Res. 2013 Jul 16;46(7):1487-96. doi: 10.1021/ar300282r. Epub 2013 Jan 7. Acc Chem Res. 2013. PMID: 23294335 Free PMC article. Review.
-
Artificial beta-sheets: chemical models of beta-sheets.Curr Opin Chem Biol. 2008 Dec;12(6):722-9. doi: 10.1016/j.cbpa.2008.08.009. Epub 2008 Sep 5. Curr Opin Chem Biol. 2008. PMID: 18775794 Free PMC article. Review.
Cited by
-
The supramolecular chemistry of β-sheets.J Am Chem Soc. 2013 Apr 17;135(15):5477-92. doi: 10.1021/ja3088407. Epub 2013 Apr 2. J Am Chem Soc. 2013. PMID: 23548073 Free PMC article. Review.
-
Synthesis and conformational analysis of bicyclic extended dipeptide surrogates.J Org Chem. 2010 Aug 6;75(15):5113-25. doi: 10.1021/jo1008433. J Org Chem. 2010. PMID: 20593836 Free PMC article.
-
Synthesis and binding studies of two new macrocyclic receptors for the stereoselective recognition of dipeptides.Beilstein J Org Chem. 2010 Jan 19;6:5. doi: 10.3762/bjoc.6.5. Beilstein J Org Chem. 2010. PMID: 20485587 Free PMC article.
-
Exploring beta-sheet structure and interactions with chemical model systems.Acc Chem Res. 2008 Oct;41(10):1319-30. doi: 10.1021/ar800064f. Epub 2008 Sep 18. Acc Chem Res. 2008. PMID: 18798654 Free PMC article.
-
A new artificial beta-sheet that dimerizes through parallel beta-sheet interactions.Org Lett. 2009 Feb 19;11(4):1003-6. doi: 10.1021/ol802993v. Org Lett. 2009. PMID: 19173616 Free PMC article.
References
-
- Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. Nature. 2005;435:773–778. - PMC - PubMed
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, MacFarlane HT, Madsen AØ, Riekel C, Eisenberg D. Nature. 2007;447:453–457. - PubMed
-
- Syud FA, Stanger HE, Gellman SH. J Am Chem Soc. 2001;123:8667–8677. 1. - PubMed
- Russell SJ, Blandl T, Skelton NJ, Cochran AG. J Am Chem Soc. 2003;125:388–395. - PubMed
- Nowick JS, Brower JO. J Am Chem Soc. 2003;125:876–877. - PubMed
- Ciani B, Jourdan M, Searle MS. J Am Chem Soc. 2003;125:9038–9047. - PubMed
- Aemissegger A, Krautler V, van Gunsteren WF, Hilvert D. J Am Chem Soc. 2005;127:2929–2936. - PubMed
- Hughes RM, Waters ML. J Am Chem Soc. 2005;127:6518–6519. - PubMed
- Mahalakshmi R, Raghothama S, Balaram P. J Am Chem Soc. 2006;128:1125–1138. - PubMed
- Hughes RM, Waters ML. J Am Chem Soc. 2006;128:13586–13591. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

