DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct
- PMID: 17920341
- PMCID: PMC2185714
- DOI: 10.1016/j.dnarep.2007.08.005
DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct
Abstract
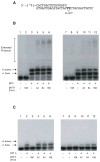
DNA polymerase theta (pol theta) is a nuclear A-family DNA polymerase encoded by the POLQ gene in vertebrate cells. The biochemical properties of pol theta and of Polq-defective mice have suggested that pol theta participates in DNA damage tolerance. For example, pol theta was previously found to be proficient not only in incorporation of a nucleotide opposite a thymine glycol or an abasic site, but also extends a polynucleotide chain efficiently from the base opposite the lesion. We carried out experiments to determine whether this ability to extend from non-standard termini is a more general property of the enzyme. Pol theta extended relatively efficiently from matched termini as well as termini with A:G, A:T and A:C mismatches, with less descrimination than a well-studied A-family DNA polymerase, exonuclease-free pol I from E. coli. Although pol theta was unable to, by itself, bypass a cyclobutane pyrimidine dimer or a (6-4) photoproduct, it could perform some extension from primers with bases placed across from these lesions. When pol theta was combined with DNA polymerase iota, an enzyme that can insert a base opposite a UV-induced (6-4) photoproduct, complete bypass of a (6-4) photoproduct was possible. These data show that in addition to its ability to insert nucleotides opposite some DNA lesions, pol theta is proficient at extension of unpaired termini. These results show the potential of pol theta to act as an extender after incorporation of nucleotides by other DNA polymerases, and aid in understanding the role of pol theta in somatic mutagenesis and genome instability.
Figures
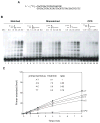
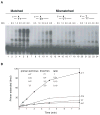
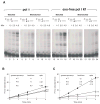
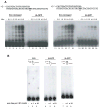

Similar articles
-
Sequence context-dependent replication of DNA templates containing UV-induced lesions by human DNA polymerase iota.DNA Repair (Amst). 2003 Sep 18;2(9):991-1006. doi: 10.1016/s1568-7864(03)00094-6. DNA Repair (Amst). 2003. PMID: 12967656
-
Misinsertion and bypass of thymine-thymine dimers by human DNA polymerase iota.EMBO J. 2000 Oct 2;19(19):5259-66. doi: 10.1093/emboj/19.19.5259. EMBO J. 2000. PMID: 11013228 Free PMC article.
-
Lesion bypass activity of DNA polymerase θ (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts.J Mol Biol. 2011 Jan 21;405(3):642-52. doi: 10.1016/j.jmb.2010.10.041. Epub 2010 Nov 2. J Mol Biol. 2011. PMID: 21050863 Free PMC article.
-
Human mitochondrial DNA polymerase γ exhibits potential for bypass and mutagenesis at UV-induced cyclobutane thymine dimers.J Biol Chem. 2012 Mar 16;287(12):9222-9. doi: 10.1074/jbc.M111.306852. Epub 2011 Dec 21. J Biol Chem. 2012. PMID: 22194617 Free PMC article.
-
DNA polymerase θ (POLQ), double-strand break repair, and cancer.DNA Repair (Amst). 2016 Aug;44:22-32. doi: 10.1016/j.dnarep.2016.05.003. Epub 2016 May 14. DNA Repair (Amst). 2016. PMID: 27264557 Free PMC article. Review.
Cited by
-
DNA Polymerase θ: A Cancer Drug Target with Reverse Transcriptase Activity.Genes (Basel). 2021 Jul 27;12(8):1146. doi: 10.3390/genes12081146. Genes (Basel). 2021. PMID: 34440316 Free PMC article. Review.
-
The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases.Cell Res. 2008 Jan;18(1):148-61. doi: 10.1038/cr.2008.4. Cell Res. 2008. PMID: 18166979 Free PMC article. Review.
-
Translesion DNA synthesis and mutagenesis in eukaryotes.Cold Spring Harb Perspect Biol. 2013 Mar 1;5(3):a012708. doi: 10.1101/cshperspect.a012708. Cold Spring Harb Perspect Biol. 2013. PMID: 23457261 Free PMC article. Review.
-
Expression and Structural Analyses of Human DNA Polymerase θ (POLQ).Methods Enzymol. 2017;592:103-121. doi: 10.1016/bs.mie.2017.03.026. Epub 2017 May 24. Methods Enzymol. 2017. PMID: 28668117 Free PMC article.
-
Promiscuous DNA synthesis by human DNA polymerase θ.Nucleic Acids Res. 2012 Mar;40(6):2611-22. doi: 10.1093/nar/gkr1102. Epub 2011 Dec 1. Nucleic Acids Res. 2012. PMID: 22135286 Free PMC article.
References
-
- Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. - PubMed
-
- Hübscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. - PubMed
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2. ASM Press; Washington, D.C: 2006.
-
- Vaisman A, Lehmann AR, Woodgate R. DNA polymerasesη and ι. Adv Protein Chem. 2004;69:205–228. - PubMed
-
- Plosky BS, Woodgate R. Switching from high-fidelity replicases to low-fidelity lesion-bypass polymerases. Curr Opin Genet Dev. 2004;14:113–119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

