Alkaline methanolysis of lipid extracts extends shotgun lipidomics analyses to the low-abundance regime of cellular sphingolipids
- PMID: 17920553
- PMCID: PMC2131739
- DOI: 10.1016/j.ab.2007.08.019
Alkaline methanolysis of lipid extracts extends shotgun lipidomics analyses to the low-abundance regime of cellular sphingolipids
Abstract
Sphingolipids that contain a sphingoid base are composed of hundreds to thousands of distinct compounds, many of which serve as lipid regulators of biological functions. The global analysis of the large number of low-abundance sphingolipid molecular species has been hampered in many cases by the sphingolipid molecular species being overwhelmed by the quantity of other classes of lipid (e.g., glycerophospholipid) molecular species present, thereby imposing severe restrictions on the dynamic range of their measurement using shotgun lipidomics. Herein, we developed a facile approach in which the sphingolipids of cellular extracts were dramatically enriched by direct alkaline methanolysis of lipid extracts followed by extraction to remove the large majority of other endogenous lipid classes. Through direct infusion of the resultant enriched solution, we identified and quantitated a variety of very-low-abundance sphingolipid classes (e.g., sphingosine, psychosine, and lysosphingomyelin) and molecular species (e.g., sphingomyelin) using electrospray ionization mass spectrometry (i.e., shotgun sphingolipidomics). Accordingly, through utilization of these facile enrichment techniques, direct penetrance into the sphingolipidomes has been greatly extended, facilitating new insights into their metabolism and signaling functions in biological systems.
Figures
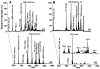


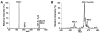



References
-
- Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J Lipid Res. 2003;44:1071–1079. - PubMed
-
- Lagarde M, Geloen A, Record M, Vance D, Spener F. Lipidomics is emerging. Biochim Biophys Acta. 2003;1634:61. - PubMed
-
- Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. - PubMed
-
- Han X, Gross RW. Shotgun lipidomics: Electrospray ionization mass spectrometric analysis and quantitation of the cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005;24:367–412. - PubMed
-
- Walker JM, Krey JF, Chen JS, Vefring E, Jahnsen JA, Bradshaw H, Huang SM. Targeted lipidomics: fatty acid amides and pain modulation. Prostaglandins Other Lipid Mediat. 2005;77:35–45. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

