Epigenetics of antigen-receptor gene assembly
- PMID: 17920858
- PMCID: PMC2151926
- DOI: 10.1016/j.gde.2007.08.006
Epigenetics of antigen-receptor gene assembly
Abstract
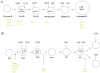
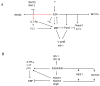
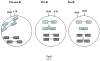
The antigen receptor genes are organized into distinct DNA elements that encode the variable (V), diversity (D) and joining (J) regions. It is now well established that the rearrangement of antigen receptor genes is regulated by developmental-specific modulation of chromatin structure. Further studies involving statistical mechanics should provide physical insight into the physical mechanisms that underlie the association of antigen receptor gene segments.
Figures
Similar articles
-
Chromatin topology and the regulation of antigen receptor assembly.Annu Rev Immunol. 2012;30:337-56. doi: 10.1146/annurev-immunol-020711-075003. Epub 2012 Jan 3. Annu Rev Immunol. 2012. PMID: 22224771 Review.
-
Epigenetic regulation of antigen receptor gene rearrangement.Curr Opin Immunol. 2011 Apr;23(2):171-7. doi: 10.1016/j.coi.2010.12.008. Epub 2011 Jan 7. Curr Opin Immunol. 2011. PMID: 21216580 Free PMC article. Review.
-
Chromatin architecture and the generation of antigen receptor diversity.Cell. 2009 Aug 7;138(3):435-48. doi: 10.1016/j.cell.2009.07.016. Cell. 2009. PMID: 19665968 Free PMC article. Review.
-
Mechanisms that control antigen receptor variable region gene assembly.Semin Immunol. 1994 Jun;6(3):185-96. doi: 10.1006/smim.1994.1024. Semin Immunol. 1994. PMID: 7948958 Review.
-
Accessibility and the developmental regulation of V(D)J recombination.Semin Immunol. 1997 Jun;9(3):161-70. doi: 10.1006/smim.1997.0066. Semin Immunol. 1997. PMID: 9200327 Review.
Cited by
-
T-cell receptor Vβ repertoire of CD8+ T-lymphocyte subpopulations in cutaneous leishmaniasis patients from the state of Rio de Janeiro, Brazil.Mem Inst Oswaldo Cruz. 2015 Aug;110(5):596-605. doi: 10.1590/0074-02760150039. Epub 2015 Jun 24. Mem Inst Oswaldo Cruz. 2015. PMID: 26107186 Free PMC article.
-
E protein binding at the Tcra enhancer promotes Tcra repertoire diversity.Front Immunol. 2023 Jul 6;14:1188738. doi: 10.3389/fimmu.2023.1188738. eCollection 2023. Front Immunol. 2023. PMID: 37483636 Free PMC article.
-
Constant domain-regulated antibody catalysis.J Biol Chem. 2012 Oct 19;287(43):36096-104. doi: 10.1074/jbc.M112.401075. Epub 2012 Sep 4. J Biol Chem. 2012. PMID: 22948159 Free PMC article.
-
Genetic variation stimulated by epigenetic modification.PLoS One. 2008;3(12):e4075. doi: 10.1371/journal.pone.0004075. Epub 2008 Dec 30. PLoS One. 2008. PMID: 19115012 Free PMC article.
References
-
- Hesslein DG, Schatz DG. Factors and forces controlling V(D)J recombination. Adv Immunol. 2001;78:169–232. - PubMed
-
- Mostoslavsky R, Alt FW, Bassing CH. Chromatin dynamics and locus accessibility in the immune system. Nat Immunol. 2003;4:603–606. - PubMed
-
- Jung D, Gillourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain gene. Annual Rev Immunol. 2006;24:541–570. - PubMed
-
- Nussenzweig MC, Shaw AC, Sinn E, Danner DB, Hohnes HC, Morse HC, III, Leder P. Allelic exclusion in transgenic mice that express the membrane form of Ig M. Science. 1988;236:816–819. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

