Genomic and biochemical studies demonstrating the absence of an alkane-producing phenotype in Vibrio furnissii M1
- PMID: 17921268
- PMCID: PMC2168193
- DOI: 10.1128/AEM.01785-07
Genomic and biochemical studies demonstrating the absence of an alkane-producing phenotype in Vibrio furnissii M1
Abstract
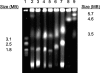
Vibrio furnissii M1 was recently reported to biosynthesize n-alkanes when grown on biopolymers, sugars, or organic acids (M. O. Park, J. Bacteriol. 187:1426-1429, 2005). In the present study, V. furnissii M1 was subjected to genomic analysis and studied biochemically. The sequence of the 16S rRNA gene and repetitive PCR showed that V. furnissii M1 was not identical to other V. furnissii strains tested, but the level of relatedness was consistent with its assignment as a V. furnissii strain. Pulsed-field gel electrophoresis showed chromosomal bands at approximately 3.2 and 1.8 Mb, similar to other Vibrio strains. Complete genomic DNA from V. furnissii M1 was sequenced with 21-fold coverage. Alkane biosynthetic and degradation genes could not be identified. Moreover, V. furnissii M1 did not produce demonstrable levels of n-alkanes in vivo or in vitro. In vivo experiments were conducted by growing V. furnissii M1 under different conditions, extracting with solvent, and analyzing extracts by gas chromatography-mass spectrometry. A highly sensitive assay was used for in vitro experiments with cell extracts and [(14)C]hexadecanol. The data are consistent with the present strain being a V. furnissii with properties similar to those previously described but lacking the alkane-producing phenotype. V. furnissii ATCC 35016, also reported to biosynthesize alkanes, was found in the present study not to produce alkanes.
Figures



Similar articles
-
New pathway for long-chain n-alkane synthesis via 1-alcohol in Vibrio furnissii M1.J Bacteriol. 2005 Feb;187(4):1426-9. doi: 10.1128/JB.187.4.1426-1429.2005. J Bacteriol. 2005. PMID: 15687207 Free PMC article.
-
Development of a simple and practical method of discrimination between Vibrio furnissii and V. fluvialis based on single-nucleotide polymorphisms of 16S rRNA genes observed in V. furnissii but not in V. fluvialis.J Microbiol Methods. 2018 Jan;144:22-28. doi: 10.1016/j.mimet.2017.10.014. Epub 2017 Oct 27. J Microbiol Methods. 2018. PMID: 29111399
-
Production of alternatives to fuel oil from organic waste by the alkane-producing bacterium, Vibrio furnissii M1.J Appl Microbiol. 2005;98(2):324-31. doi: 10.1111/j.1365-2672.2004.02454.x. J Appl Microbiol. 2005. PMID: 15659187
-
Phenotypic and genotypic characterization of Vibrio viscosus sp. nov. and Vibrio wodanis sp. nov. isolated from Atlantic salmon (Salmo salar) with 'winter ulcer'.Int J Syst Evol Microbiol. 2000 Mar;50 Pt 2:427-450. doi: 10.1099/00207713-50-2-427. Int J Syst Evol Microbiol. 2000. PMID: 10758846
-
Post-Genomic Analysis of Members of the Family Vibrionaceae.Microbiol Spectr. 2015 Oct;3(5):10.1128/microbiolspec.VE-0009-2014. doi: 10.1128/microbiolspec.VE-0009-2014. Microbiol Spectr. 2015. PMID: 26542048 Free PMC article. Review.
Cited by
-
Genes involved in long-chain alkene biosynthesis in Micrococcus luteus.Appl Environ Microbiol. 2010 Feb;76(4):1212-23. doi: 10.1128/AEM.02312-09. Epub 2009 Dec 28. Appl Environ Microbiol. 2010. PMID: 20038703 Free PMC article.
-
Widespread head-to-head hydrocarbon biosynthesis in bacteria and role of OleA.Appl Environ Microbiol. 2010 Jun;76(12):3850-62. doi: 10.1128/AEM.00436-10. Epub 2010 Apr 23. Appl Environ Microbiol. 2010. PMID: 20418421 Free PMC article.
-
C29 olefinic hydrocarbons biosynthesized by Arthrobacter species.Appl Environ Microbiol. 2009 Mar;75(6):1774-7. doi: 10.1128/AEM.02547-08. Epub 2009 Jan 23. Appl Environ Microbiol. 2009. PMID: 19168653 Free PMC article.
-
Bacterial microcompartments: their properties and paradoxes.Bioessays. 2008 Nov;30(11-12):1084-95. doi: 10.1002/bies.20830. Bioessays. 2008. PMID: 18937343 Free PMC article. Review.
-
Synthetic biology and biomass conversion: a match made in heaven?J R Soc Interface. 2009 Aug 6;6 Suppl 4(Suppl 4):S547-58. doi: 10.1098/rsif.2008.0527.focus. Epub 2009 May 19. J R Soc Interface. 2009. PMID: 19454530 Free PMC article. Review.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Alvarez, H. M., and A. Steinbuchel. 2002. Triacylglycerols in prokaryotic microorganisms. Appl. Microbiol. Biotechnol. 60:367-376. - PubMed
-
- Bassler, B. L., C. Yu, Y. C. Lee, and S. Roseman. 1991. Chitin utilization by marine bacteria. Degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J. Biol. Chem. 266:24276-24286. - PubMed
-
- Bligh, E. G., and W. J. Dyer. 1959. A rapid method for total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. - PubMed
-
- Bobik, T. A. 2006. Polyhedral organelles compartmenting bacterial metabolic processes. Appl. Microbiol. Biotechnol. 70:517-525. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

