Molecular analysis of the diversity of sulfate-reducing and sulfur-oxidizing prokaryotes in the environment, using aprA as functional marker gene
- PMID: 17921272
- PMCID: PMC2168068
- DOI: 10.1128/AEM.01272-07
Molecular analysis of the diversity of sulfate-reducing and sulfur-oxidizing prokaryotes in the environment, using aprA as functional marker gene
Abstract
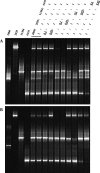
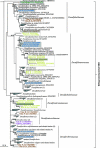
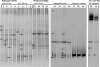
The dissimilatory adenosine-5'-phosphosulfate reductase is a key enzyme of the microbial sulfate reduction and sulfur oxidation processes. Because the alpha- and beta-subunit-encoding genes, aprBA, are highly conserved among sulfate-reducing and sulfur-oxidizing prokaryotes, they are most suitable for molecular profiling of the microbial community structure of the sulfur cycle in environment. In this study, a new aprA gene-targeting assay using a combination of PCR and denaturing gradient gel electrophoresis is presented. The screening of sulfate-reducing and sulfur-oxidizing reference strains as well as the analyses of environmental DNA from diverse habitats (e.g., microbial mats, invertebrate tissue, marine and estuarine sediments, and filtered hydrothermal water) by the new primer pair revealed an improved microbial diversity coverage and less-pronounced template-to-PCR product bias in direct comparison to those of the previously published primer set (B. Deplancke, K. R. Hristova, H. A. Oakley, V. J. McCracken, R. Aminov, R. I. Mackie, and H. R. Gaskins, Appl. Environ. Microbiol. 66:2166-2174, 2000). The concomitant molecular detection of sulfate-reducing and sulfur-oxidizing prokaryotes was confirmed. The new assay was applied in comparison with the 16S rRNA gene-based analysis to investigate the microbial diversity of the sulfur cycle in sediment, seawater, and manganese crust samples from four study sites in the area of the Lesser Antilles volcanic arc, Caribbean Sea (Caribflux project). The aprA gene-based approach revealed putative sulfur-oxidizing Alphaproteobacteria of chemolithoheterotrophic lifestyle to have been abundant in the nonhydrothermal sediment and water column. In contrast, the sulfur-based microbial community that inhabited the surface of the volcanic manganese crust was more complex, consisting predominantly of putative chemolithoautotrophic sulfur oxidizers of the Betaproteobacteria and Gammaproteobacteria.
Figures


References
-
- Arakawa, S., T. Sato, Y. Yoshida, R. Usami, and C. Kato. 2006. Comparison of the microbial diversity in cold-seep sediments from different depths in the Nankai Trough. J. Gen. Appl. Microbiol. 52:47-54. - PubMed
-
- Bagwell, C. E., X. Liu, L. Wu, and J. Zhou. 2006. Effects of legacy nuclear waste on the compositional diversity and distributions of sulfate-reducing bacteria in a terrestrial subsurface aquifer. FEMS Microbiol. Ecol. 55:424-431. - PubMed
-
- Bahr, M., B. C. Crump, V. Klepac-Ceraj, A. Teske, M. L. Sogin, and J. E. Hobbie. 2005. Molecular characterization of sulfate-reducing bacteria in a New England salt marsh. Environ. Microbiol. 7:1175-1185. - PubMed
-
- Brüser, T., P. N. L. Lens, and H. G. Trüper. 2000. The biological sulfur cycle, p. 47-86. In P. N. L. Lens and L. H. Pol (ed.), Environmental technologies to treat sulfur pollution. IWA Publishing, London, United Kingdom.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

