Chromosomal and symbiotic relationships of rhizobia nodulating Medicago truncatula and M. laciniata
- PMID: 17921275
- PMCID: PMC2168047
- DOI: 10.1128/AEM.01046-07
Chromosomal and symbiotic relationships of rhizobia nodulating Medicago truncatula and M. laciniata
Abstract
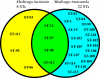
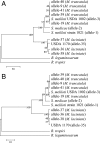
Multilocus sequence typing (MLST) is a sequence-based method used to characterize bacterial genomes. This method was used to examine the genetic structure of Medicago-nodulating rhizobia at the Amra site, which is located in an arid region of Tunisia. Here the annual medics Medicago laciniata and M. truncatula are part of the natural flora. The goal of this study was to identify whether distinct chromosomal groups of rhizobia nodulate M. laciniata because of its restricted requirement for specific rhizobia. The MLST analysis involved determination of sequence variation in 10 chromosomal loci of 74 isolates each of M. laciniata and M. truncatula. M. truncatula was used as a control trap host, because unlike M. laciniata, it has relatively unrestricted rhizobial requirements. Allelic diversity among the plasmid nodC alleles in the isolates was also determined. The 148 isolates were placed into 26 chromosomal sequence types (STs), only 3 of which had been identified previously. The rhizobia of M. laciniata were shown to be part of the general Medicago-nodulating population in the soil because 99.95% of the isolates had chromosomal genotypes similar to those recovered from M. truncatula. However, the isolates recovered from M. laciniata were less diverse than those recovered from M. truncatula, and they also harbored an unusual nodC allele. This could perhaps be best explained by horizontal transfer of the different nodC alleles among members of the Medicago-nodulating rhizobial population at the field site. Evidence indicating a history of lateral transfer of rhizobial symbiotic genes across distinct chromosomal backgrounds is provided.
Figures


References
-
- Badri, Y., K. Zribi, M. Badri, T. Huguet, and M. E. Aouani. 2003. Sinorhizobium meliloti nodulates Medicago laciniata in Tunisian soils. Czech J. Genet. Plant Breed. 39(Special Issue):178-183.
-
- Ballard, R. A., and N. Charman. 2000. Nodulation and growth of pasture legumes with naturalised soil rhizobia. 1. Annual Medicago spp. Aust. J. Exp. Agric. 40:939-948.
-
- Barran, L. R., E. S. P. Bromfield, and D. C. W. Brown. 2002. Identification and cloning of the bacterial nodulation specificity gene in the Sinorhizobium meliloti-Medicago laciniata symbiosis. Can. J. Microbiol. 48:765-771. - PubMed
-
- Biondi, E. G., E. Pilli, E. Giuntini, M. L. Roumiantseva, E. E. Andronov, O. P. Onichtchouk, O. N. Kurchak, B. V. Simarov, N. I. Dzyubenko, A. Mengoni, and M. Bazzicalupo. 2003. Genetic relationship of Sinorhizobium meliloti and Sinorhizobium medicae strains isolated from Caucasian region. FEMS Microbiol. 220:207-213. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

