Sequence characterization and comparative analysis of three plasmids isolated from environmental Vibrio spp
- PMID: 17921277
- PMCID: PMC2168046
- DOI: 10.1128/AEM.01577-07
Sequence characterization and comparative analysis of three plasmids isolated from environmental Vibrio spp
Abstract
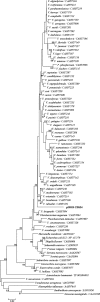
The horizontal transfer of genes by mobile genetic elements such as plasmids and phages can accelerate genome diversification of Vibrio spp., affecting their physiology, pathogenicity, and ecological character. In this study, sequence analysis of three plasmids from Vibrio spp. previously isolated from salt marsh sediment revealed the remarkable diversity of these elements. Plasmids p0908 (81.4 kb), p23023 (52.5 kb), and p09022 (31.0 kb) had a predicted 99, 64, and 32 protein-coding sequences and G+C contents of 49.2%, 44.7%, and 42.4%, respectively. A phylogenetic tree based on concatenation of the host 16S rRNA and rpoA nucleotide sequences indicated p23023 and p09022 were isolated from strains most closely related to V. mediterranei and V. campbellii, respectively, while the host of p0908 forms a clade with V. fluvialis and V. furnissii. Many predicted proteins had amino acid identities to proteins of previously characterized phages and plasmids (24 to 94%). Predicted proteins with similarity to chromosomally encoded proteins included RecA, a nucleoid-associated protein (NdpA), a type IV helicase (UvrD), and multiple hypothetical proteins. Plasmid p0908 had striking similarity to enterobacteria phage P1, sharing genetic organization and amino acid identity for 23 predicted proteins. This study provides evidence of genetic exchange between Vibrio plasmids, phages, and chromosomes among diverse Vibrio spp.
Figures


Similar articles
-
Vibrio ishigakensis sp. nov., in Halioticoli clade isolated from seawater in Okinawa coral reef area, Japan.Syst Appl Microbiol. 2016 Jul;39(5):330-5. doi: 10.1016/j.syapm.2016.04.002. Epub 2016 May 24. Syst Appl Microbiol. 2016. PMID: 27262360
-
Advanced Microbial Taxonomy Combined with Genome-Based-Approaches Reveals that Vibrio astriarenae sp. nov., an Agarolytic Marine Bacterium, Forms a New Clade in Vibrionaceae.PLoS One. 2015 Aug 27;10(8):e0136279. doi: 10.1371/journal.pone.0136279. eCollection 2015. PLoS One. 2015. PMID: 26313925 Free PMC article.
-
Vibrio olivae sp. nov., isolated from Spanish-style green-olive fermentations.Int J Syst Evol Microbiol. 2015 Jun;65(Pt 6):1895-1901. doi: 10.1099/ijs.0.000196. Epub 2015 Mar 19. Int J Syst Evol Microbiol. 2015. PMID: 25792368
-
Vibrio rhizosphaerae sp. nov., a red-pigmented bacterium that antagonizes phytopathogenic bacteria.Int J Syst Evol Microbiol. 2007 Oct;57(Pt 10):2241-2246. doi: 10.1099/ijs.0.65017-0. Int J Syst Evol Microbiol. 2007. PMID: 17911290
-
What do we know about plasmids carried by members of the Acinetobacter genus?World J Microbiol Biotechnol. 2020 Jul 13;36(8):109. doi: 10.1007/s11274-020-02890-7. World J Microbiol Biotechnol. 2020. PMID: 32656745 Review.
Cited by
-
Genome diversification within a clonal population of pandemic Vibrio parahaemolyticus seems to depend on the life circumstances of each individual bacteria.BMC Genomics. 2015 Mar 13;16(1):176. doi: 10.1186/s12864-015-1385-8. BMC Genomics. 2015. PMID: 25880192 Free PMC article.
-
Endogenous and Foreign Nucleoid-Associated Proteins of Bacteria: Occurrence, Interactions and Effects on Mobile Genetic Elements and Host's Biology.Comput Struct Biotechnol J. 2019 Jun 14;17:746-756. doi: 10.1016/j.csbj.2019.06.010. eCollection 2019. Comput Struct Biotechnol J. 2019. PMID: 31303979 Free PMC article. Review.
-
VCre/VloxP and SCre/SloxP: new site-specific recombination systems for genome engineering.Nucleic Acids Res. 2011 Apr;39(8):e49. doi: 10.1093/nar/gkq1280. Epub 2011 Feb 1. Nucleic Acids Res. 2011. PMID: 21288882 Free PMC article.
-
Insights into the environmental reservoir of pathogenic Vibrio parahaemolyticus using comparative genomics.Front Microbiol. 2015 Mar 24;6:204. doi: 10.3389/fmicb.2015.00204. eCollection 2015. Front Microbiol. 2015. PMID: 25852665 Free PMC article.
-
Infaunal burrows are enrichment zones for Vibrio parahaemolyticus.Appl Environ Microbiol. 2011 Jun;77(11):3703-14. doi: 10.1128/AEM.02897-10. Epub 2011 Apr 8. Appl Environ Microbiol. 2011. PMID: 21478307 Free PMC article.
References
-
- Agron, P. G., P. Sobecky, and G. L. Andersen. 2002. Establishment of uncharacterized plasmids in Escherichia coli by in vitro transposition. FEMS Microbiol. Lett. 217:249-254. - PubMed
-
- Ahmed, A. M., S. Shinoda, and T. Shimamoto. 2005. A variant type of Vibrio cholerae SXT element in a multidrug-resistant strain of Vibrio fluvialis. FEMS Microbiol. Lett. 242:241-247. - PubMed
-
- Balding, C., S. A. Bromley, R. W. Pickup, and J. R. Saunders. 2005. Diversity of phage integrases in Enterobacteriaceae: development of markers for environmental analysis of temperate phages. Environ. Microbiol. 7:1558-1567. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

