Functional analysis of the M.HpyAIV DNA methyltransferase of Helicobacter pylori
- PMID: 17921292
- PMCID: PMC2168601
- DOI: 10.1128/JB.00108-07
Functional analysis of the M.HpyAIV DNA methyltransferase of Helicobacter pylori
Abstract
A large number of genes encoding restriction-modification (R-M) systems are found in the genome of the human pathogen Helicobacter pylori. R-M genes comprise approximately 10% of the strain-specific genes, but the relevance of having such an abundance of these genes is not clear. The type II methyltransferase (MTase) M.HpyAIV, which recognizes GANTC sites, was present in 60% of the H. pylori strains analyzed, whereof 69% were resistant to restriction enzyme digestion, which indicated the presence of an active MTase. H. pylori strains with an inactive M.HpyAIV phenotype contained deletions in regions of homopolymers within the gene, which resulted in premature translational stops, suggesting that M.HpyAIV may be subjected to phase variation by a slipped-strand mechanism. An M.HpyAIV gene mutant was constructed by insertional mutagenesis, and this mutant showed the same viability and ability to induce interleukin-8 in epithelial cells as the wild type in vitro but had, as expected, lost the ability to protect its self-DNA from digestion by a cognate restriction enzyme. The M.HpyAIV from H. pylori strain 26695 was overexpressed in Escherichia coli, and the protein was purified and was able to bind to DNA and protect GANTC sites from digestion in vitro. A bioinformatic analysis of the number of GANTC sites located in predicted regulatory regions of H. pylori strains 26695 and J99 resulted in a number of candidate genes. katA, a selected candidate gene, was further analyzed by quantitative real-time reverse transcription-PCR and shown to be significantly down-regulated in the M.HpyAIV gene mutant compared to the wild-type strain. This demonstrates the influence of M.HpyAIV methylation in gene expression.
Figures
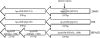
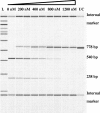

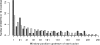
References
-
- Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. - PubMed
-
- Appelmelk, B. J., S. L. Martin, M. A. Monteiro, C. A. Clayton, A. A. McColm, P. Zheng, T. Verboom, J. J. Maaskant, D. H. van den Eijnden, C. H. Hokke, M. B. Perry, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 1999. Phase variation in Helicobacter pylori lipopolysaccharide due to changes in the lengths of poly(C) tracts in α3-fucosyltransferase genes. Infect. Immun. 67:5361-5366. - PMC - PubMed
-
- Aro, P., T. Storskrubb, J. Ronkainen, E. Bolling-Sternevald, L. Engstrand, M. Vieth, M. Stolte, N. J. Talley, and L. Agreus. 2006. Peptic ulcer disease in a general adult population: the Kalixanda study: a random population-based study. Am. J. Epidemiol. 163:1025-1034. - PubMed
-
- Atherton, J. C., P. Cao, R. M. J. Peek, M. K. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771-17777. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

