Role of the PsbI protein in photosystem II assembly and repair in the cyanobacterium Synechocystis sp. PCC 6803
- PMID: 17921338
- PMCID: PMC2151680
- DOI: 10.1104/pp.107.107805
Role of the PsbI protein in photosystem II assembly and repair in the cyanobacterium Synechocystis sp. PCC 6803
Abstract
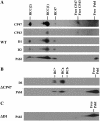
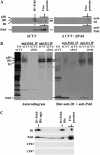
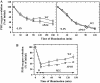
The involvement of the PsbI protein in the assembly and repair of the photosystem II (PSII) complex has been studied in the cyanobacterium Synechocystis sp. PCC 6803. Analysis of PSII complexes in the wild-type strain showed that the PsbI protein was present in dimeric and monomeric core complexes, core complexes lacking CP43, and in reaction center complexes containing D1, D2, and cytochrome b-559. In addition, immunoprecipitation experiments and the use of a histidine-tagged derivative of PsbI have revealed the presence in the thylakoid membrane of assembly complexes containing PsbI and either the precursor or mature forms of D1. Analysis of PSII assembly in the psbI deletion mutant and in strains lacking PsbI together with other PSII subunits showed that PsbI was not required for formation of PSII reaction center complexes or core complexes, although levels of unassembled D1 were reduced in its absence. However, loss of PsbI led to a dramatic destabilization of CP43 binding within monomeric and dimeric PSII core complexes. Despite the close structural relationship between D1 and PsbI in the PSII complex, PsbI turned over much slower than D1, whereas high light-induced turnover of D1 was accelerated in the absence of PsbI. Overall, our results suggest that PsbI is an early assembly partner for D1 and that it plays a functional role in stabilizing the binding of CP43 in the PSII holoenzyme.
Figures


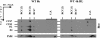

Similar articles
-
The Psb27 assembly factor binds to the CP43 complex of photosystem II in the cyanobacterium Synechocystis sp. PCC 6803.Plant Physiol. 2012 Jan;158(1):476-86. doi: 10.1104/pp.111.184184. Epub 2011 Nov 15. Plant Physiol. 2012. PMID: 22086423 Free PMC article.
-
The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803.J Biol Chem. 2008 Aug 15;283(33):22390-9. doi: 10.1074/jbc.M801917200. Epub 2008 Jun 12. J Biol Chem. 2008. PMID: 18550538
-
CyanoP is Involved in the Early Steps of Photosystem II Assembly in the Cyanobacterium Synechocystis sp. PCC 6803.Plant Cell Physiol. 2016 Sep;57(9):1921-31. doi: 10.1093/pcp/pcw115. Epub 2016 Jul 7. Plant Cell Physiol. 2016. PMID: 27388341
-
Recent advances in understanding the assembly and repair of photosystem II.Ann Bot. 2010 Jul;106(1):1-16. doi: 10.1093/aob/mcq059. Epub 2010 Mar 25. Ann Bot. 2010. PMID: 20338950 Free PMC article. Review.
-
Assembly and Repair of Photosystem II in Chlamydomonas reinhardtii.Plants (Basel). 2024 Mar 12;13(6):811. doi: 10.3390/plants13060811. Plants (Basel). 2024. PMID: 38592843 Free PMC article. Review.
Cited by
-
Complexome profiling on the Chlamydomonas lpa2 mutant reveals insights into PSII biogenesis and new PSII associated proteins.J Exp Bot. 2022 Jan 5;73(1):245-262. doi: 10.1093/jxb/erab390. J Exp Bot. 2022. PMID: 34436580 Free PMC article.
-
Importance of the cyanobacterial Gun4 protein for chlorophyll metabolism and assembly of photosynthetic complexes.J Biol Chem. 2008 Sep 19;283(38):25794-802. doi: 10.1074/jbc.M803787200. Epub 2008 Jul 14. J Biol Chem. 2008. PMID: 18625715 Free PMC article.
-
Photosystem II Assembly from Scratch.Front Plant Sci. 2016 Jan 12;6:1234. doi: 10.3389/fpls.2015.01234. eCollection 2015. Front Plant Sci. 2016. PMID: 26793213 Free PMC article. No abstract available.
-
Characterization of a Synechocystis double mutant lacking the photosystem II assembly factors YCF48 and Sll0933.Planta. 2013 Feb;237(2):471-80. doi: 10.1007/s00425-012-1720-0. Epub 2012 Jul 31. Planta. 2013. PMID: 22847023
-
How to build functional thylakoid membranes: from plastid transcription to protein complex assembly.Planta. 2013 Feb;237(2):413-28. doi: 10.1007/s00425-012-1752-5. Epub 2012 Sep 14. Planta. 2013. PMID: 22976450 Free PMC article. Review.
References
-
- Barber J (2006) Photosystem II: an enzyme of global significance. Biochem Soc Trans 34 619–631 - PubMed
-
- Bumba L, Tichý M, Dobáková M, Komenda J, Vácha F (2005) Localization of the PsbH subunit in photosystem II from the Synechocystis 6803 using the His-tagged Ni-NTA nanogold labeling. J Struct Biol 152 28–35 - PubMed
-
- Burke E, Barik S (2003) Megaprimer PCR: application in mutagenesis and gene fusion. Methods Mol Biol 226 525–532 - PubMed
-
- Eaton-Rye JJ, Vermaas WFJ (1991) Oligonucleotide-directed mutagenesis of psbB, the gene encoding CP47, employing a deletion mutant strain of the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 17 1165–1177 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

