Regulatory interactions between two actin nucleators, Spire and Cappuccino
- PMID: 17923532
- PMCID: PMC2064741
- DOI: 10.1083/jcb.200706196
Regulatory interactions between two actin nucleators, Spire and Cappuccino
Abstract
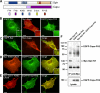
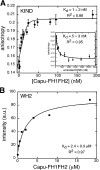
Spire and Cappuccino are actin nucleation factors that are required to establish the polarity of Drosophila melanogaster oocytes. Their mutant phenotypes are nearly identical, and the proteins interact biochemically. We find that the interaction between Spire and Cappuccino family proteins is conserved across metazoan phyla and is mediated by binding of the formin homology 2 (FH2) domain from Cappuccino (or its mammalian homologue formin-2) to the kinase noncatalytic C-lobe domain (KIND) from Spire. In vitro, the KIND domain is a monomeric folded domain. Two KIND monomers bind each FH2 dimer with nanomolar affinity and strongly inhibit actin nucleation by the FH2 domain. In contrast, formation of the Spire-Cappuccino complex enhances actin nucleation by Spire. In Drosophila oocytes, Spire localizes to the cortex early in oogenesis and disappears around stage 10b, coincident with the onset of cytoplasmic streaming.
Figures




Similar articles
-
Structure and function of the interacting domains of Spire and Fmn-family formins.Proc Natl Acad Sci U S A. 2011 Jul 19;108(29):11884-9. doi: 10.1073/pnas.1105703108. Epub 2011 Jul 5. Proc Natl Acad Sci U S A. 2011. PMID: 21730168 Free PMC article.
-
Spire stimulates nucleation by Cappuccino and binds both ends of actin filaments.Mol Biol Cell. 2020 Feb 15;31(4):273-286. doi: 10.1091/mbc.E19-09-0550. Epub 2019 Dec 26. Mol Biol Cell. 2020. PMID: 31877067 Free PMC article.
-
Interaction between microtubules and the Drosophila formin Cappuccino and its effect on actin assembly.J Biol Chem. 2014 Feb 14;289(7):4395-404. doi: 10.1074/jbc.M113.499921. Epub 2013 Dec 20. J Biol Chem. 2014. PMID: 24362037 Free PMC article.
-
Cellular functions of the Spir actin-nucleation factors.Trends Cell Biol. 2006 Sep;16(9):477-83. doi: 10.1016/j.tcb.2006.07.005. Epub 2006 Aug 9. Trends Cell Biol. 2006. PMID: 16901698 Review.
-
The WH2 Domain and Actin Nucleation: Necessary but Insufficient.Trends Biochem Sci. 2016 Jun;41(6):478-490. doi: 10.1016/j.tibs.2016.03.004. Epub 2016 Apr 5. Trends Biochem Sci. 2016. PMID: 27068179 Free PMC article. Review.
Cited by
-
Formins at a glance.J Cell Sci. 2013 Jan 1;126(Pt 1):1-7. doi: 10.1242/jcs.107250. J Cell Sci. 2013. PMID: 23516326 Free PMC article. Review. No abstract available.
-
Autoinhibition of the formin Cappuccino in the absence of canonical autoinhibitory domains.Mol Biol Cell. 2012 Oct;23(19):3801-13. doi: 10.1091/mbc.E12-04-0288. Epub 2012 Aug 8. Mol Biol Cell. 2012. PMID: 22875983 Free PMC article.
-
Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1.J Cell Biol. 2010 Jun 28;189(7):1087-96. doi: 10.1083/jcb.201001016. Epub 2010 Jun 21. J Cell Biol. 2010. PMID: 20566685 Free PMC article.
-
Dual control of formin-nucleated actin assembly by the chromatin and ER in mouse oocytes.Curr Biol. 2022 Sep 26;32(18):4013-4024.e6. doi: 10.1016/j.cub.2022.07.058. Epub 2022 Aug 17. Curr Biol. 2022. PMID: 35981539 Free PMC article.
-
Coordinated recruitment of Spir actin nucleators and myosin V motors to Rab11 vesicle membranes.Elife. 2016 Sep 13;5:e17523. doi: 10.7554/eLife.17523. Elife. 2016. PMID: 27623148 Free PMC article.
References
-
- Ciccarelli, F.D., P. Bork, and E. Kerkhoff. 2003. The KIND module: a putative signalling domain evolved from the C lobe of the protein kinase fold. Trends Biochem. Sci. 28:349–352. - PubMed
-
- Clark, I., E. Giniger, H. Ruohola-Baker, L.Y. Jan, and Y.N. Jan. 1994. Transient posterior localization of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Curr. Biol. 4:289–300. - PubMed
-
- Cooper, J.A., S.B. Walker, and T.D. Pollard. 1983. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell Motil. 4:253–262. - PubMed
-
- Emmons, S., H. Phan, J. Calley, W. Chen, B. James, and L. Manseau. 1995. Cappuccino, a Drosophila maternal effect gene required for polarity of the egg and embryo, is related to the vertebrate limb deformity locus. Genes Dev. 9:2482–2494. - PubMed
-
- Harris, E.S., F. Li, and H.N. Higgs. 2004. The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J. Biol. Chem. 279:20076–20087. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

