3'-O-modified nucleotides as reversible terminators for pyrosequencing
- PMID: 17923668
- PMCID: PMC2034218
- DOI: 10.1073/pnas.0707495104
3'-O-modified nucleotides as reversible terminators for pyrosequencing
Abstract
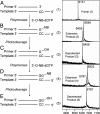
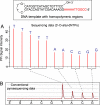
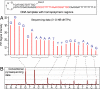
Pyrosequencing is a method used to sequence DNA by detecting the pyrophosphate (PPi) group that is generated when a nucleotide is incorporated into the growing DNA strand in polymerase reaction. However, this method has an inherent difficulty in accurately deciphering the homopolymeric regions of the DNA templates. We report here the development of a method to solve this problem by using nucleotide reversible terminators. These nucleotide analogues are modified with a reversible chemical moiety capping the 3'-OH group to temporarily terminate the polymerase reaction. In this way, only one nucleotide is incorporated into the growing DNA strand even in homopolymeric regions. After detection of the PPi for sequence determination, the 3'-OH of the primer extension products is regenerated through different deprotection methods. Using an allyl or a 2-nitrobenzyl group as the reversible moiety to cap the 3'-OH of the four nucleotides, we have synthesized two sets of 3'-O-modified nucleotides, 3'-O-allyl-dNTPs and 3'-O-(2-nitrobenzyl)-dNTPs as reversible terminators for pyrosequencing. The capping moiety on the 3'-OH of the DNA extension product is efficiently removed after PPi detection by either a chemical method or photolysis. To sequence DNA, templates containing homopolymeric regions are immobilized on Sepharose beads, and then extension-signal detection-deprotection cycles are conducted by using the nucleotide reversible terminators on the DNA beads to unambiguously decipher the sequence of DNA templates. Our results establish that this reversible-terminator-pyrosequencing approach can be potentially developed into a powerful methodology to accurately determine DNA sequences.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

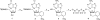


Similar articles
-
An integrated system for DNA sequencing by synthesis using novel nucleotide analogues.Acc Chem Res. 2010 Apr 20;43(4):551-63. doi: 10.1021/ar900255c. Acc Chem Res. 2010. PMID: 20121268 Free PMC article.
-
Four-color DNA sequencing with 3'-O-modified nucleotide reversible terminators and chemically cleavable fluorescent dideoxynucleotides.Proc Natl Acad Sci U S A. 2008 Jul 8;105(27):9145-50. doi: 10.1073/pnas.0804023105. Epub 2008 Jun 30. Proc Natl Acad Sci U S A. 2008. PMID: 18591653 Free PMC article.
-
Four-color DNA sequencing by synthesis using cleavable fluorescent nucleotide reversible terminators.Proc Natl Acad Sci U S A. 2006 Dec 26;103(52):19635-40. doi: 10.1073/pnas.0609513103. Epub 2006 Dec 14. Proc Natl Acad Sci U S A. 2006. PMID: 17170132 Free PMC article.
-
Translesion DNA synthesis: polymerase response to altered nucleotides.Cancer Surv. 1985;4(3):493-516. Cancer Surv. 1985. PMID: 2825983 Review.
-
The history and advances of reversible terminators used in new generations of sequencing technology.Genomics Proteomics Bioinformatics. 2013 Feb;11(1):34-40. doi: 10.1016/j.gpb.2013.01.003. Epub 2013 Jan 23. Genomics Proteomics Bioinformatics. 2013. PMID: 23414612 Free PMC article. Review.
Cited by
-
DNA and RNA analyses in detection of genetic predisposition to cancer.Hered Cancer Clin Pract. 2012 Dec 4;10(1):17. doi: 10.1186/1897-4287-10-17. Hered Cancer Clin Pract. 2012. PMID: 23206658 Free PMC article.
-
An integrated system for DNA sequencing by synthesis using novel nucleotide analogues.Acc Chem Res. 2010 Apr 20;43(4):551-63. doi: 10.1021/ar900255c. Acc Chem Res. 2010. PMID: 20121268 Free PMC article.
-
A new method to synthesize multiple gRNA libraries and functional mapping of mammalian H3K4me3 regions.Nucleic Acids Res. 2023 May 22;51(9):e50. doi: 10.1093/nar/gkad198. Nucleic Acids Res. 2023. PMID: 36938898 Free PMC article.
-
Four-color DNA sequencing with 3'-O-modified nucleotide reversible terminators and chemically cleavable fluorescent dideoxynucleotides.Proc Natl Acad Sci U S A. 2008 Jul 8;105(27):9145-50. doi: 10.1073/pnas.0804023105. Epub 2008 Jun 30. Proc Natl Acad Sci U S A. 2008. PMID: 18591653 Free PMC article.
-
Mini review: Enzyme-based DNA synthesis and selective retrieval for data storage.Comput Struct Biotechnol J. 2021 Apr 25;19:2468-2476. doi: 10.1016/j.csbj.2021.04.057. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34025937 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

