T-bet's ability to regulate individual target genes requires the conserved T-box domain to recruit histone methyltransferase activity and a separate family member-specific transactivation domain
- PMID: 17923685
- PMCID: PMC2169399
- DOI: 10.1128/MCB.01615-07
T-bet's ability to regulate individual target genes requires the conserved T-box domain to recruit histone methyltransferase activity and a separate family member-specific transactivation domain
Abstract
Appropriate cellular differentiation and specification rely upon the ability of key developmental transcription factors to precisely establish gene expression patterns. These transcription factors often regulate epigenetic events. However, it has been unclear whether this is the only role that they play in functionally regulating developmental gene expression pathways or whether they also participate in downstream transactivation events at the same promoter. The T-box transcription factor family is important in cellular specification events in many developmental systems, and determining the molecular mechanisms by which this family regulates gene expression networks warrants attention. Here, we examine the mechanism by which T-bet, a critical T-box protein in the immune system, influences transcription. T-bet is both necessary and sufficient to induce permissive histone H3-K4 dimethyl modifications at the CXCR3 and IFN-gamma promoters. A T-bet structure-function analysis revealed that the conserved T-box domain, with a small C-terminal portion, is required for recruiting histone methyltransferase activity to promoters. Interestingly, this function is conserved in the T-box family and is necessary, but not sufficient, to induce transcription, with an independent transactivation activity also required. The requirement for two separable functional activities may ultimately contribute to the stringent role for T-box proteins in establishing specific developmental gene expression pathways.
Figures




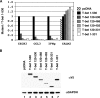


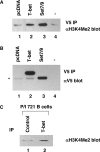
Similar articles
-
An essential interaction between T-box proteins and histone-modifying enzymes.Epigenetics. 2009 Feb 16;4(2):85-8. doi: 10.4161/epi.4.2.8111. Epigenetics. 2009. PMID: 19384057 Review.
-
Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression.Genes Dev. 2008 Nov 1;22(21):2980-93. doi: 10.1101/gad.1689708. Genes Dev. 2008. PMID: 18981476 Free PMC article.
-
Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression.Mol Cell. 2010 Nov 24;40(4):594-605. doi: 10.1016/j.molcel.2010.10.028. Mol Cell. 2010. PMID: 21095589 Free PMC article.
-
Opposing roles of STAT4 and Dnmt3a in Th1 gene regulation.J Immunol. 2013 Jul 15;191(2):902-11. doi: 10.4049/jimmunol.1203229. Epub 2013 Jun 14. J Immunol. 2013. PMID: 23772023 Free PMC article.
-
Molecular mechanisms by which T-bet regulates T-helper cell commitment.Immunol Rev. 2010 Nov;238(1):233-46. doi: 10.1111/j.1600-065X.2010.00952.x. Immunol Rev. 2010. PMID: 20969596 Free PMC article. Review.
Cited by
-
Ezh2 regulates transcriptional and posttranslational expression of T-bet and promotes Th1 cell responses mediating aplastic anemia in mice.J Immunol. 2014 Jun 1;192(11):5012-22. doi: 10.4049/jimmunol.1302943. Epub 2014 Apr 23. J Immunol. 2014. PMID: 24760151 Free PMC article.
-
Transcription factor-dependent chromatin remodeling of Il18r1 during Th1 and Th2 differentiation.J Immunol. 2008 Sep 1;181(5):3346-52. doi: 10.4049/jimmunol.181.5.3346. J Immunol. 2008. PMID: 18714006 Free PMC article.
-
T-bet-dependent S1P5 expression in NK cells promotes egress from lymph nodes and bone marrow.J Exp Med. 2009 Oct 26;206(11):2469-81. doi: 10.1084/jem.20090525. Epub 2009 Oct 6. J Exp Med. 2009. PMID: 19808259 Free PMC article.
-
IL-2-inducible T-cell kinase modulates TH2-mediated allergic airway inflammation by suppressing IFN-γ in naive CD4+ T cells.J Allergy Clin Immunol. 2013 Oct;132(4):811-20.e1-5. doi: 10.1016/j.jaci.2013.04.033. Epub 2013 Jun 12. J Allergy Clin Immunol. 2013. PMID: 23768572 Free PMC article.
-
Transcriptomic Alterations in Lung Adenocarcinoma Unveil New Mechanisms Targeted by the TBX2 Subfamily of Tumor Suppressor Genes.Front Oncol. 2018 Oct 30;8:482. doi: 10.3389/fonc.2018.00482. eCollection 2018. Front Oncol. 2018. PMID: 30425966 Free PMC article.
References
-
- Afkarin, M., J. R. Sedy, J. Yang, N. G. Jacobson, N. Cereb, S. Y. Yang, T. L. Murphy, and K. M. Murphy. 2002. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat. Immunol. 3:549-557. - PubMed
-
- Agarwal, S., and A. Rao. 1998. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity 9:765-775. - PubMed
-
- Ansel, K. M., I. Djuretic, B. Tanasa, and A. Rao. 2006. Regulation of Th2 differentiation and IL4 locus accessibility. Annu. Rev. Immunol. 24:607-656. - PubMed
-
- Avni, O., D. Lee, F. Macian, S. J. Szabo, L. H. Glimcher, and A. Rao. 2002. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat. Immunol. 3:643-651. - PubMed
-
- Baala, L., S. Briault, H. C. Etchevers, F. Laumonnier, A. Natiq, J. Amiel, N. Boddaert, C. Picard, A. Sbiti, A. Asermouh, T. Attie-Bitach, F. Encha-Razavi, A. Munnich, A. Sefiani, and S. Lyonnet. 2007. Homozygous silencing of T-box transcription factor EOMES leads to microcephaly with polymicrogyria and corpus callosum agenesis. Nat. Genet. 39:454-456. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
