dE2F2-independent rescue of proliferation in cells lacking an activator dE2F1
- PMID: 17923695
- PMCID: PMC2169406
- DOI: 10.1128/MCB.01068-07
dE2F2-independent rescue of proliferation in cells lacking an activator dE2F1
Abstract
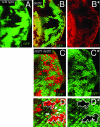
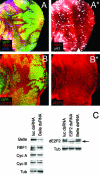
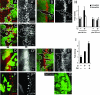
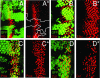
In Drosophila melanogaster, the loss of activator de2f1 leads to a severe reduction in cell proliferation and repression of E2F targets. To date, the only known way to rescue the proliferation block in de2f1 mutants was through the inactivation of dE2F2. This suggests that dE2F2 provides a major contribution to the de2f1 mutant phenotype. Here, we report that in mosaic animals, in addition to de2f2, the loss of a DEAD box protein Belle (Bel) also rescues proliferation of de2f1 mutant cells. Surprisingly, the rescue occurs in a dE2F2-independent manner since the loss of Bel does not relieve dE2F2-mediated repression. In the eye disc, bel mutant cells fail to undergo a G1 arrest in the morphogenetic furrow, delay photoreceptor recruitment and differentiation, and show a reduction of the transcription factor Ci155. The down-regulation of Ci155 is important since it is sufficient to partially rescue proliferation of de2f1 mutant cells. Thus, mutation of bel relieves the dE2F2-mediated cell cycle arrest in de2f1 mutant cells through a novel Ci155-dependent mechanism without functional inactivation of the dE2F2 repressor.
Figures



References
-
- Arora, K., M. B. O'Connor, and R. Warrior. 1996. BMP signaling in Drosophila embryogenesis. Ann. N. Y. Acad. Sci. 785:80-97. - PubMed
-
- Baonza, A., and M. Freeman. 2005. Control of cell proliferation in the Drosophila eye by Notch signaling. Dev. Cell 8:529-539. - PubMed
-
- Cayirlioglu, P., P. C. Bonnette, M. R. Dickson, and R. J. Duronio. 2001. Drosophila E2f2 promotes the conversion from genomic DNA replication to gene amplification in ovarian follicle cells. Development 128:5085-5098. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
