Sumoylation modulates the assembly and activity of the pre-mRNA 3' processing complex
- PMID: 17923699
- PMCID: PMC2169417
- DOI: 10.1128/MCB.01186-07
Sumoylation modulates the assembly and activity of the pre-mRNA 3' processing complex
Abstract
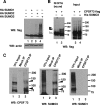
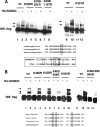
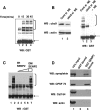
Eukaryotic pre-mRNA 3'-end formation is catalyzed by a complex set of factors that must be intricately regulated. In this study, we have discovered a novel role for the small ubiquitin-like modifier SUMO in the regulation of mammalian 3'-end processing. We identified symplekin, a factor involved in complex assembly, and CPSF-73, an endonuclease, as SUMO modification substrates. The major sites of sumoylation in symplekin and CPSF-73 were determined and found to be highly conserved across species. A sumoylation-deficient mutant was defective in rescuing cell viability in symplekin small interfering RNA (siRNA)-treated cells, supporting the importance of this modification in symplekin function. We also analyzed the involvement of sumoylation in 3'-end processing by altering the sumoylation status of nuclear extracts. This was done by the addition of a SUMO protease, which we show interacts with both symplekin and CPSF-73, or by siRNA-mediated depletion of ubc9, the SUMO E2-conjugating enzyme. Both treatments resulted in a marked inhibition of processing. The assembly of a functional polyadenylation complex was also impaired by the SUMO protease. Our identification of two key polyadenylation factors as SUMO targets and of the role of SUMO in enhancing the assembly and activity of the 3'-end-processing complex together reveal an important function for SUMO in the processing of mRNA precursors.
Figures




References
-
- Barabino, S. M., and W. Keller. 1999. Last but not least: regulated poly(A) tail formation. Cell 99:9-11. - PubMed
-
- Barnard, D. C., K. Ryan, J. L. Manley, and J. D. Richter. 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119:641-651. - PubMed
-
- Best, J. L., S. Ganiatsas, S. Agarwal, A. Changou, P. Salomoni, O. Shirihai, P. B. Meluh, P. P. Pandolfi, and L. I. Zon. 2002. SUMO-1 protease-1 regulates gene transcription through PML. Mol. Cell 10:843-855. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
