Transcription alters chromosomal locations of cohesin in Saccharomyces cerevisiae
- PMID: 17923700
- PMCID: PMC2169412
- DOI: 10.1128/MCB.01007-07
Transcription alters chromosomal locations of cohesin in Saccharomyces cerevisiae
Abstract
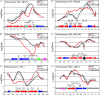
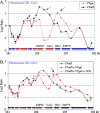
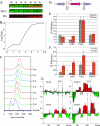
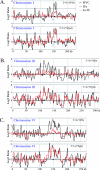
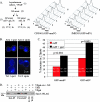
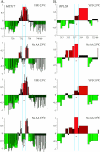
In eukaryotic cells, cohesion between sister chromatids allows chromosomes to biorient on the metaphase plate and holds them together until they separate into daughter cells during mitosis. Cohesion is mediated by the cohesin protein complex. Although the association of this complex with particular regions of the genome is highly reproducible, it is unclear what distinguishes a chromosomal region for cohesin association. Since one of the primary locations of cohesin is intergenic regions between converging transcription units, we explored the relationship between transcription and cohesin localization. Chromatin immunoprecipitation followed by hybridization to a microarray (ChIP chip) indicated that transcript elongation into cohesin association sites results in the local disassociation of cohesin. Once transcription is halted, cohesin can reassociate with its original sites, independent of DNA replication and the cohesin loading factor Scc2, although cohesin association with chromosomes in G2/M is not functional for cohesion. A computer program was developed to systematically identify differences between two ChIP chip data sets. Our results are consistent with a model for cohesin association in which (i) a portion of cohesin can be dynamically loaded and unloaded to accommodate transcription and (ii) the cohesin complex has preferences for features of chromatin that are a reflection of the local transcriptional status. Taken together, our results suggest that cohesion may be degraded by transcription.
Figures
References
-
- Antoniacci, L. M., and R. V. Skibbens. 2006. Sister-chromatid telomere cohesion is nonredundant and resists both spindle forces and telomere motility. Curr. Biol. 16:902-906. - PubMed
-
- Blat, Y., and N. Kleckner. 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98:249-259. - PubMed
-
- Bohlander, S. K., R. Espinosa III, M. M. Le Beau, J. D. Rowley, and M. O. Diaz. 1992. A method for the rapid sequence-independent amplification of microdissected chromosomal material. Genomics 13:1322-1324. - PubMed
-
- Ciosk, R., M. Shirayama, A. Shevchenko, T. Tanaka, A. Toth, and K. Nasmyth. 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5:243-254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
