A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes
- PMID: 17924331
- PMCID: PMC2265654
- DOI: 10.1086/521032
A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes
Abstract
Mutation screening of the breast and ovarian cancer-predisposition genes BRCA1 and BRCA2 is becoming an increasingly important part of clinical practice. Classification of rare nontruncating sequence variants in these genes is problematic, because it is not known whether these subtle changes alter function sufficiently to predispose cells to cancer development. Using data from the Myriad Genetic Laboratories database of nearly 70,000 full-sequence tests, we assessed the clinical significance of 1,433 sequence variants of unknown significance (VUSs) in the BRCA genes. Three independent measures were employed in the assessment: co-occurrence in trans of a VUS with known deleterious mutations; detailed analysis, by logistic regression, of personal and family history of cancer in VUS-carrying probands; and, in a subset of probands, an analysis of cosegregation with disease in pedigrees. For each of these factors, a likelihood ratio was computed under the hypothesis that the VUSs were equivalent to an "average" deleterious mutation, compared with neutral, with respect to risk. The likelihood ratios derived from each component were combined to provide an overall assessment for each VUS. A total of 133 VUSs had odds of at least 100 : 1 in favor of neutrality with respect to risk, whereas 43 had odds of at least 20 : 1 in favor of being deleterious. VUSs with evidence in favor of causality were those that were predicted to affect splicing, fell at positions that are highly conserved among BRCA orthologs, and were more likely to be located in specific domains of the proteins. In addition to their utility for improved genetics counseling of patients and their families, the global assessment reported here will be invaluable for validation of functional assays, structural models, and in silico analyses.
Figures
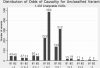
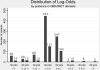
References
Web Resources
-
- Align GVGD, http://agvgd.iarc.fr/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BRCA1 and BRCA2)
References
-
- Burke W, Daly M, Garber J, Botkin J, Kahn MJ, Lynch P, McTiernan A, Offit K, Perlman J, Petersen G, et al (1997) Recommendations for follow-up care of individuals with an inherited predisposition to cancer: II. BRCA1 and BRCA2—Cancer Genetics Studies Consortium. JAMA 277:997–1003 10.1001/jama.277.12.997 - DOI - PubMed
-
- Lucci-Cordisco E, Boccuto L, Neri G, Genuardi M (2006) The use of microsatellite instability, immunohistochemistry and other variables in determining the clinical significance of MLH1 and MSH2 unclassified variants in Lynch syndrome. Cancer Biomark 2:11–27 - PubMed
-
- Billack B, Monteiro AN (2004) Methods to classify BRCA1 variants of uncertain clinical significance: the more the merrier. Cancer Biol Ther 3:458–459 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

