Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy
- PMID: 17924332
- PMCID: PMC2265642
- DOI: 10.1086/521986
Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy
Abstract
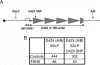
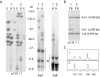
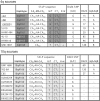
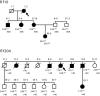
Autosomal dominant facioscapulohumeral muscular dystrophy (FSHD) is mainly characterized by progressive wasting and weakness of the facial, shoulder, and upper-arm muscles. FSHD is caused by contraction of the macrosatellite repeat D4Z4 on chromosome 4q35. The D4Z4 repeat is very polymorphic in length, and D4Z4 rearrangements occur almost exclusively via intrachromosomal gene conversions. Several disease mechanisms have been proposed, but none of these models can comprehensively explain FSHD, because repeat contraction alone is not sufficient to cause disease. Almost-identical D4Z4-repeat arrays have been identified on chromosome 10q26 and on two equally common chromosome 4 variants, 4qA and 4qB. Yet only repeat contractions of D4Z4 on chromosome 4qA cause FSHD; contractions on the other chromosomes are nonpathogenic. We hypothesized that allele-specific sequence differences among 4qA, 4qB, and 10q alleles underlie the 4qA specificity of FSHD. Sequence variations between these alleles have been described before, but the extent and significance of these variations proximal to, within, and distal to D4Z4 have not been studied in detail. We examined additional sequence variations in the FSHD locus, including a relatively stable simple sequence-length polymorphism proximal to D4Z4, a single-nucleotide polymorphism (SNP) within D4Z4, and the A/B variation distal to D4Z4. On the basis of these polymorphisms, we demonstrate that the subtelomeric domain of chromosome 4q can be subdivided into nine distinct haplotypes, of which three carry the distal 4qA variation. Interestingly, we show that repeat contractions in two of the nine haplotypes, one of which is a 4qA haplotype, are not associated with FSHD. We also show that each of these haplotypes has its unique sequence signature, and we propose that specific SNPs in the disease haplotype are essential for the development of FSHD.
Figures


References
Web Resources
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for SSLP sequences [accession numbers AF117653, AL845259, AY028079, BX649463, BX294170, BX005259, and AL954635])
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FSHD1A) - PubMed
References
-
- Padberg GW (1982) Facioscapulohumeral disease. Leiden University, Leiden
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

