Fine mapping versus replication in whole-genome association studies
- PMID: 17924341
- PMCID: PMC2265650
- DOI: 10.1086/521952
Fine mapping versus replication in whole-genome association studies
Abstract
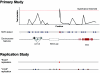
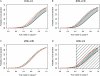
Association replication studies have a poor track record and, even when successful, often claim association with different markers, alleles, and phenotypes than those reported in the primary study. It is unknown whether these outcomes reflect genuine associations or false-positive results. A greater understanding of these observations is essential for genomewide association (GWA) studies, since they have the potential to identify multiple new associations that that will require external validation. Theoretically, a repeat association with precisely the same variant in an independent sample is the gold standard for replication, but testing additional variants is commonplace in replication studies. Finding different associated SNPs within the same gene or region as that originally identified is often reported as confirmatory evidence. Here, we compare the probability of replicating a gene or region under two commonly used marker-selection strategies: an "exact" approach that involves only the originally significant markers and a "local" approach that involves both the originally significant markers and others in the same region. When a region of high intermarker linkage disequilibrium is tested to replicate an initial finding that is only weak association with disease, the local approach is a good strategy. Otherwise, the most powerful and efficient strategy for replication involves testing only the initially identified variants. Association with a marker other than that originally identified can occur frequently, even in the presence of real effects in a low-powered replication study, and instances of such association increase as the number of included variants increases. Our results provide a basis for the design and interpretation of GWA replication studies and point to the importance of a clear distinction between fine mapping and replication after GWA.
Figures




References
Web Resource
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for IL23R and Crohn disease)
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

