Waves of genomic hitchhikers shed light on the evolution of gamebirds (Aves: Galliformes)
- PMID: 17925025
- PMCID: PMC2169234
- DOI: 10.1186/1471-2148-7-190
Waves of genomic hitchhikers shed light on the evolution of gamebirds (Aves: Galliformes)
Abstract
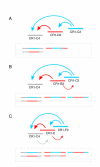
Background: The phylogenetic tree of Galliformes (gamebirds, including megapodes, currassows, guinea fowl, New and Old World quails, chicken, pheasants, grouse, and turkeys) has been considerably remodeled over the last decades as new data and analytical methods became available. Analyzing presence/absence patterns of retroposed elements avoids the problems of homoplastic characters inherent in other methodologies. In gamebirds, chicken repeats 1 (CR1) are the most prevalent retroposed elements, but little is known about the activity of their various subtypes over time. Ascertaining the fixation patterns of CR1 elements would help unravel the phylogeny of gamebirds and other poorly resolved avian clades.
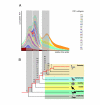
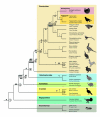
Results: We analyzed 1,978 nested CR1 elements and developed a multidimensional approach taking advantage of their transposition in transposition character (TinT) to characterize the fixation patterns of all 22 known chicken CR1 subtypes. The presence/absence patterns of those elements that were active at different periods of gamebird evolution provided evidence for a clade (Cracidae + (Numididae + (Odontophoridae + Phasianidae))) not including Megapodiidae; and for Rollulus as the sister taxon of the other analyzed Phasianidae. Genomic trace sequences of the turkey genome further demonstrated that the endangered African Congo Peafowl (Afropavo congensis) is the sister taxon of the Asian Peafowl (Pavo), rejecting other predominantly morphology-based groupings, and that phasianids are monophyletic, including the sister taxa Tetraoninae and Meleagridinae.
Conclusion: The TinT information concerning relative fixation times of CR1 subtypes enabled us to efficiently investigate gamebird phylogeny and to reconstruct an unambiguous tree topology. This method should provide a useful tool for investigations in other taxonomic groups as well.
Figures
References
-
- Fain MG, Houde P. Parallel radiations in the primary clades of birds. Evolution. 2004;58:2558–2573. - PubMed
-
- Crowe TM. The evolution of guineafowl: (Galliformes, Phasianidae, Numidinae): taxonomy, phylogeny, speciation and biogeography. Ann S Afr Mus. 1978;76:43–136.
-
- Gutiérrez RJ, Zink RM, Yang SY. Genic variation, systematic biogeographic relationships of some galliform birds. The Auk. 1983;100:33–47.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

