Toxicogenomic analysis incorporating operon-transcriptional coupling and toxicant concentration-expression response: analysis of MX-treated Salmonella
- PMID: 17925033
- PMCID: PMC2225428
- DOI: 10.1186/1471-2105-8-378
Toxicogenomic analysis incorporating operon-transcriptional coupling and toxicant concentration-expression response: analysis of MX-treated Salmonella
Abstract
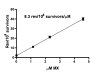
Background: Deficiencies in microarray technology cause unwanted variation in the hybridization signal, obscuring the true measurements of intracellular transcript levels. Here we describe a general method that can improve microarray analysis of toxicant-exposed cells that uses the intrinsic power of transcriptional coupling and toxicant concentration-expression response data. To illustrate this approach, we characterized changes in global gene expression induced in Salmonella typhimurium TA100 by 3-chloro-4-(dichloromethyl)-5-hydroxy-2(5H)-furanone (MX), the primary mutagen in chlorinated drinking water. We used the co-expression of genes within an operon and the monotonic increases or decreases in gene expression relative to increasing toxicant concentration to augment our identification of differentially expressed genes beyond Bayesian-t analysis.
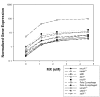
Results: Operon analysis increased the number of altered genes by 95% from the list identified by a Bayesian t-test of control to the highest concentration of MX. Monotonic analysis added 46% more genes. A functional analysis of the resulting 448 differentially expressed genes yielded functional changes beyond what would be expected from only the mutagenic properties of MX. In addition to gene-expression changes in DNA-damage response, MX induced changes in expression of genes involved in membrane transport and porphyrin metabolism, among other biological processes. The disruption of porphyrin metabolism might be attributable to the structural similarity of MX, which is a chlorinated furanone, to ligands indigenous to the porphyrin metabolism pathway. Interestingly, our results indicate that the lexA regulon in Salmonella, which partially mediates the response to DNA damage, may contain only 60% of the genes present in this regulon in E. coli. In addition, nanH was found to be highly induced by MX and contains a putative lexA regulatory motif in its regulatory region, suggesting that it may be regulated by lexA.
Conclusion: Operon and monotonic analyses improved the determination of differentially expressed genes beyond that of Bayesian-t analysis, showing that MX alters cellular metabolism involving pathways other than DNA damage. Because co-expression of similarly functioning genes also occurs in eukaryotes, this method has general applicability for improving analysis of toxicogenomic data.
Figures


Similar articles
-
Mutation spectra of the drinking water mutagen 3-chloro-4-methyl-5-hydroxy-2(5H)-furanone (MCF) in Salmonella TA100 and TA104: comparison to MX.Environ Mol Mutagen. 2000;35(2):106-13. Environ Mol Mutagen. 2000. PMID: 10712744
-
Mutagen structure and transcriptional response: induction of distinct transcriptional profiles in Salmonella TA100 by the drinking-water mutagen MX and its homologues.Environ Mol Mutagen. 2010 Jan;51(1):69-79. doi: 10.1002/em.20512. Environ Mol Mutagen. 2010. PMID: 19598237
-
Mutation spectra in Salmonella of analogues of MX: implications of chemical structure for mutational mechanisms.Mutat Res. 2000 Sep 20;453(1):51-65. doi: 10.1016/s0027-5107(00)00084-1. Mutat Res. 2000. PMID: 11006412
-
Carcinogenicity of the chlorination disinfection by-product MX.J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2005;23(2):163-214. doi: 10.1080/10590500500234988. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2005. PMID: 16291527 Review.
-
Mutagenic compounds in chlorinated pulp bleaching waters and drinking waters.IARC Sci Publ. 1990;(104):333-40. IARC Sci Publ. 1990. PMID: 2228130 Review.

