X chromosome inactivation during Drosophila spermatogenesis
- PMID: 17927450
- PMCID: PMC2001211
- DOI: 10.1371/journal.pbio.0050273
X chromosome inactivation during Drosophila spermatogenesis
Abstract
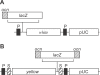
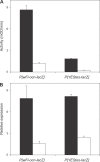
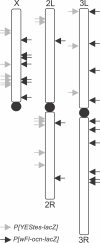
Genes with male- and testis-enriched expression are under-represented on the Drosophila melanogaster X chromosome. There is also an excess of retrotransposed genes, many of which are expressed in testis, that have "escaped" the X chromosome and moved to the autosomes. It has been proposed that inactivation of the X chromosome during spermatogenesis contributes to these patterns: genes with a beneficial function late in spermatogenesis should be selectively favored to be autosomal in order to avoid inactivation. However, conclusive evidence for X inactivation in the male germline has been lacking. To test for such inactivation, we used a transgenic construct in which expression of a lacZ reporter gene was driven by the promoter sequence of the autosomal, testis-specific ocnus gene. Autosomal insertions of this transgene showed the expected pattern of male- and testis-specific expression. X-linked insertions, in contrast, showed only very low levels of reporter gene expression. Thus, we find that X linkage inhibits the activity of a testis-specific promoter. We obtained the same result using a vector in which the transgene was flanked by chromosomal insulator sequences. These results are consistent with global inactivation of the X chromosome in the male germline and support a selective explanation for X chromosome avoidance of genes with beneficial effects late in spermatogenesis.
Conflict of interest statement
Figures

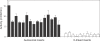
Similar articles
-
Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes.PLoS Genet. 2009 Nov;5(11):e1000731. doi: 10.1371/journal.pgen.1000731. Epub 2009 Nov 20. PLoS Genet. 2009. PMID: 19936020 Free PMC article.
-
Fine-scale analysis of X chromosome inactivation in the male germ line of Drosophila melanogaster.Mol Biol Evol. 2011 May;28(5):1561-3. doi: 10.1093/molbev/msq355. Epub 2010 Dec 30. Mol Biol Evol. 2011. PMID: 21193490
-
'Escaping' the X chromosome leads to increased gene expression in the male germline of Drosophila melanogaster.Heredity (Edinb). 2014 Feb;112(2):149-55. doi: 10.1038/hdy.2013.86. Epub 2013 Sep 11. Heredity (Edinb). 2014. PMID: 24022496 Free PMC article.
-
Doubling the rewards: testis ESTs for Drosophila gene discovery and spermatogenesis expression profile analysis.Genome Res. 2000 Dec;10(12):1841-2. doi: 10.1101/gr.169400. Genome Res. 2000. PMID: 11116081 Review. No abstract available.
-
Mammalian Sex Chromosome Structure, Gene Content, and Function in Male Fertility.Annu Rev Anim Biosci. 2019 Feb 15;7:103-124. doi: 10.1146/annurev-animal-020518-115332. Epub 2018 Nov 9. Annu Rev Anim Biosci. 2019. PMID: 30412673 Review.
Cited by
-
Demasculinization of the Anopheles gambiae X chromosome.BMC Evol Biol. 2012 May 18;12:69. doi: 10.1186/1471-2148-12-69. BMC Evol Biol. 2012. PMID: 22607633 Free PMC article.
-
Retrogenes reveal the direction of sex-chromosome evolution in mosquitoes.Genetics. 2010 Oct;186(2):763-6. doi: 10.1534/genetics.110.118794. Epub 2010 Jul 26. Genetics. 2010. PMID: 20660646 Free PMC article.
-
Lack of global meiotic sex chromosome inactivation, and paucity of tissue-specific gene expression on the Drosophila X chromosome.BMC Biol. 2011 May 4;9:29. doi: 10.1186/1741-7007-9-29. BMC Biol. 2011. PMID: 21542906 Free PMC article.
-
Effects of X-linkage and sex-biased gene expression on the rate of adaptive protein evolution in Drosophila.Mol Biol Evol. 2008 Aug;25(8):1639-50. doi: 10.1093/molbev/msn111. Epub 2008 May 13. Mol Biol Evol. 2008. PMID: 18477586 Free PMC article.
-
Functional characterization of adaptive variation within a cis-regulatory element influencing Drosophila melanogaster growth.PLoS Biol. 2018 Jan 11;16(1):e2004538. doi: 10.1371/journal.pbio.2004538. eCollection 2018 Jan. PLoS Biol. 2018. PMID: 29324742 Free PMC article.
References
-
- Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr Biol. 1996;6:149–162. - PubMed
-
- Rice WR. Evolution of the Y sex chromosome in animals. Bioscience. 1996;46:331–343.
-
- Steinemann M, Steinemann S. Common mechanisms of Y chromosome evolution. Genetica. 2000;109:105–111. - PubMed
-
- Steinemann S, Steinemann M. Biased distribution of repetitive elements: A landmark for neo-Y chromosome evolution in Drosophila miranda . Cytogenet Cell Genet. 2001;93:228–233. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

