A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae
- PMID: 17927698
- PMCID: PMC2586181
- DOI: 10.1111/j.1365-2958.2007.05966.x
A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae
Abstract
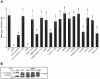
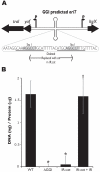
The Neisseria gonorrhoeae type IV secretion system secretes chromosomal DNA that acts in natural transformation. To examine the mechanism of DNA processing for secretion, we made mutations in the putative relaxase gene traI and used nucleases to characterize the secreted DNA. The nuclease experiments demonstrated that the secreted DNA is single-stranded and blocked at the 5' end. Mutation of traI identified Tyr93 as required for DNA secretion, while substitution of Tyr201 resulted in intermediate levels of DNA secretion. TraI exhibits features of relaxases, but also has features that are absent in previously characterized relaxases, including an HD phosphohydrolase domain and an N-terminal hydrophobic region. The HD domain residue Asp120 was required for wild-type levels of DNA secretion. Subcellular localization studies demonstrated that the TraI N-terminal region promotes membrane interaction. We propose that Tyr93 initiates DNA processing and Tyr201 is required for termination or acts in DNA binding. Disruption of an inverted-repeat sequence eliminated DNA secretion, suggesting that this sequence may serve as the origin of transfer for chromosomal DNA secretion. The TraI domain architecture, although not previously described, is present in 53 uncharacterized proteins, suggesting that the mechanism of TraI function is a widespread process for DNA donation.
Figures
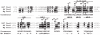






References
-
- Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23:469–472. - PubMed
-
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. - PubMed
-
- Bordo D, Argos P. Suggestions for “safe” residue substitutions in site-directed mutagenesis. J Mol Biol. 1991;217:721–729. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

