More effective purifying selection on RNA viruses than in DNA viruses
- PMID: 17928171
- PMCID: PMC2756238
- DOI: 10.1016/j.gene.2007.09.013
More effective purifying selection on RNA viruses than in DNA viruses
Abstract
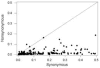
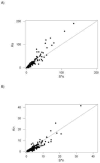
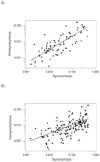
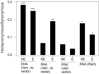
Analysis of the pattern of nucleotide diversity in 222 independent viral sequence datasets showed the prevalence of purifying selection. In spite of the higher mutation rate of RNA viruses, our analyses revealed stronger evidence of the action of purifying selection in RNA viruses than in DNA viruses. The ratio of nonsynonymous to synonymous nucleotide diversity was significantly lower in RNA viruses than in DNA viruses, indicating that nonsynonymous mutations have been removed at a greater rate (relative to the mutation rate) in the former than in the latter. Moreover, statistics that measure the occurrence of rare polymorphisms revealed significantly a greater excess of rare nonsynonymous polymorphisms in RNA viruses than in DNA viruses but no difference with respect to synonymous polymorphisms. Since rare nonsynonymous polymorphisms are likely to be undergoing the effects of purifying selection acting to eliminate them, this result implies a stronger signature of ongoing purifying selection in RNA viruses than in DNA viruses. Across datasets from both DNA viruses and RNA viruses, we found a negatively allometric relationship between nonsynonymous and synonymous nucleotide diversity; in other words, nonsynonymous nucleotide diversity increased with synonymous nucleotide diversity at a less than linear rate. These findings are most easily explained by the occurrence of slightly deleterious mutations. The fact that the negative allometry was more pronounced in RNA viruses than in DNA viruses provided additional evidence that purifying selection is more effective in the former than in the latter.
Figures
Similar articles
-
Micro-scale signature of purifying selection in Marburg virus genomes.Gene. 2007 May 1;392(1-2):266-72. doi: 10.1016/j.gene.2006.12.038. Epub 2007 Jan 24. Gene. 2007. PMID: 17306473
-
The fitness effects of synonymous mutations in DNA and RNA viruses.Mol Biol Evol. 2012 Jan;29(1):17-20. doi: 10.1093/molbev/msr179. Epub 2011 Jul 18. Mol Biol Evol. 2012. PMID: 21771719
-
Small effective population sizes and rare nonsynonymous variants in potyviruses.Virology. 2009 Oct 10;393(1):127-34. doi: 10.1016/j.virol.2009.07.016. Epub 2009 Aug 19. Virology. 2009. PMID: 19695658 Free PMC article.
-
Extensive purifying selection acting on synonymous sites in HIV-1 Group M sequences.Virol J. 2008 Dec 23;5:160. doi: 10.1186/1743-422X-5-160. Virol J. 2008. PMID: 19105834 Free PMC article. Review.
-
Evolutionary forces at work in partitiviruses.Virus Genes. 2019 Oct;55(5):563-573. doi: 10.1007/s11262-019-01680-0. Epub 2019 Jun 22. Virus Genes. 2019. PMID: 31230256 Review.
Cited by
-
Evolutionary trajectory of fish Piscine novirhabdovirus (=Viral Hemorrhagic Septicemia Virus) across its Laurentian Great Lakes history: Spatial and temporal diversification.Ecol Evol. 2020 Sep 2;10(18):9740-9775. doi: 10.1002/ece3.6611. eCollection 2020 Sep. Ecol Evol. 2020. PMID: 33005343 Free PMC article.
-
Banana bunchy top virus genetic diversity in Pakistan and association of diversity with recombination in its genomes.PLoS One. 2022 Mar 7;17(3):e0263875. doi: 10.1371/journal.pone.0263875. eCollection 2022. PLoS One. 2022. PMID: 35255085 Free PMC article.
-
Genomic and immunogenic changes of Piscine novirhabdovirus (Viral Hemorrhagic Septicemia Virus) over its evolutionary history in the Laurentian Great Lakes.PLoS One. 2021 May 28;16(5):e0232923. doi: 10.1371/journal.pone.0232923. eCollection 2021. PLoS One. 2021. PMID: 34048438 Free PMC article.
-
Molecular Analysis of Full-Length VP2 of Canine Parvovirus Reveals Antigenic Drift in CPV-2b and CPV-2c Variants in Central Chile.Animals (Basel). 2021 Aug 12;11(8):2387. doi: 10.3390/ani11082387. Animals (Basel). 2021. PMID: 34438844 Free PMC article.
-
Natural Selection Plays an Important Role in Shaping the Codon Usage of Structural Genes of the Viruses Belonging to the Coronaviridae Family.Viruses. 2020 Dec 22;13(1):3. doi: 10.3390/v13010003. Viruses. 2020. PMID: 33375017 Free PMC article.
References
-
- Agol VI. Molecular mechanisms of poliovirus variation and evolution. Curr Top Microbiol Immunol. 2006;299:211–259. - PubMed
-
- Allen TM, et al. Tat-specific CTL select for SIV escape variants during resolution of primary viremia. Nature. 2000;407:386–390. - PubMed
-
- Berlin S, Ellegren H. Fast accumulation of nonsynonymous mutations on the female-specific W chromosome in birds. J Mol Evol. 2006;62:66–72. - PubMed
-
- Chao L. Evolution of sex in RNA viruses. J Theor Biol. 1988;133:99–112. - PubMed
-
- Chao L. Evolution of sex and the molecular clock in RNA viruses. Gene. 1997;205:301–308. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

