Discrete clusters of virus-encoded micrornas are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus
- PMID: 17928340
- PMCID: PMC2168849
- DOI: 10.1128/JVI.01290-07
Discrete clusters of virus-encoded micrornas are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus
Abstract
The prevalence and importance of microRNAs (miRNAs) in viral infection are increasingly relevant. Eleven miRNAs were previously identified in human cytomegalovirus (HCMV); however, miRNA content in murine CMV (MCMV), which serves as an important in vivo model for CMV infection, has not previously been examined. We have cloned and characterized 17 novel miRNAs that originate from at least 12 precursor miRNAs in MCMV and are not homologous to HCMV miRNAs. In parallel, we applied a computational analysis, using a support vector machine approach, to identify potential precursor miRNAs in MCMV. Four of the top 10 predicted precursor sequences were cloned in this study, and the combination of computational and cloning analysis demonstrates that MCMV has the capacity to encode miRNAs clustered throughout the genome. On the basis of drug sensitivity experiments for resolving the kinetic class of expression, we show that the MCMV miRNAs are both early and late gene products. Notably, the MCMV miRNAs occur on complementary strands of the genome in specific regions, a feature which has not previously been observed for viral miRNAs. One cluster of miRNAs occurs in close proximity to the 5' splice site of the previously identified 7.2-kb stable intron, implying a variety of potential regulatory mechanisms for MCMV miRNAs.
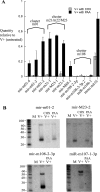
Figures




Similar articles
-
Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation.J Virol. 2007 Dec;81(24):13771-82. doi: 10.1128/JVI.01313-07. Epub 2007 Oct 17. J Virol. 2007. PMID: 17942535 Free PMC article.
-
Manipulation of Viral MicroRNAs as a Potential Antiviral Strategy for the Treatment of Cytomegalovirus Infection.Viruses. 2017 May 19;9(5):118. doi: 10.3390/v9050118. Viruses. 2017. PMID: 28534856 Free PMC article.
-
Cloning, characterization, and expression of the murine cytomegalovirus homologue of the human cytomegalovirus 28-kDa matrix phosphoprotein (UL99).Virology. 1994 Dec;205(2):417-29. doi: 10.1006/viro.1994.1662. Virology. 1994. PMID: 7975245
-
Repair of an Attenuated Low-Passage Murine Cytomegalovirus Bacterial Artificial Chromosome Identifies a Novel Spliced Gene Essential for Salivary Gland Tropism.J Virol. 2020 Oct 27;94(22):e01456-20. doi: 10.1128/JVI.01456-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32847854 Free PMC article.
-
Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target.Proc Natl Acad Sci U S A. 2012 Jan 3;109(1):279-84. doi: 10.1073/pnas.1114204109. Epub 2011 Dec 19. Proc Natl Acad Sci U S A. 2012. PMID: 22184245 Free PMC article.
Cited by
-
Integrated analysis of mRNA and viral miRNAs in the kidney of Carassius auratus gibelio response to cyprinid herpesvirus 2.Sci Rep. 2017 Oct 23;7(1):13787. doi: 10.1038/s41598-017-14217-y. Sci Rep. 2017. PMID: 29062054 Free PMC article.
-
Mapping the Human Herpesvirus 6B transcriptome.J Virol. 2021 Apr 26;95(10):e01335-20. doi: 10.1128/JVI.01335-20. Epub 2021 Feb 24. J Virol. 2021. PMID: 33627386 Free PMC article.
-
The functions of herpesvirus-encoded microRNAs.Med Microbiol Immunol. 2008 Jun;197(2):261-7. doi: 10.1007/s00430-007-0070-1. Epub 2007 Dec 18. Med Microbiol Immunol. 2008. PMID: 18087721 Free PMC article. Review.
-
Human Cytomegalovirus Host Interactions: EGFR and Host Cell Signaling Is a Point of Convergence Between Viral Infection and Functional Changes in Infected Cells.Front Microbiol. 2021 May 7;12:660901. doi: 10.3389/fmicb.2021.660901. eCollection 2021. Front Microbiol. 2021. PMID: 34025614 Free PMC article. Review.
-
MicroRNAs expressed by human cytomegalovirus.Virol J. 2020 Mar 12;17(1):34. doi: 10.1186/s12985-020-1296-4. Virol J. 2020. PMID: 32164742 Free PMC article. Review.
References
-
- Bennasser, Y., S. Y. Le, M. L. Yeung, and K. T. Jeang. 2006. MicroRNAs in human immunodeficiency virus-1 infection. Methods Mol. Biol. 342:241-253. - PubMed
-
- Bonnet, E., J. Wuyts, P. Rouze, and Y. Van de Peer. 2004. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20:2911-2917. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

