A cysteine-rich CCG domain contains a novel [4Fe-4S] cluster binding motif as deduced from studies with subunit B of heterodisulfide reductase from Methanothermobacter marburgensis
- PMID: 17929940
- PMCID: PMC3543786
- DOI: 10.1021/bi700679u
A cysteine-rich CCG domain contains a novel [4Fe-4S] cluster binding motif as deduced from studies with subunit B of heterodisulfide reductase from Methanothermobacter marburgensis
Abstract
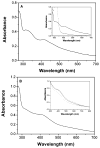
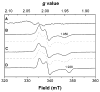
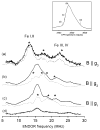
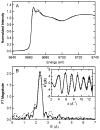
Heterodisulfide reductase (HDR) of methanogenic archaea with its active-site [4Fe-4S] cluster catalyzes the reversible reduction of the heterodisulfide (CoM-S-S-CoB) of the methanogenic coenzyme M (CoM-SH) and coenzyme B (CoB-SH). CoM-HDR, a mechanistic-based paramagnetic intermediate generated upon half-reaction of the oxidized enzyme with CoM-SH, is a novel type of [4Fe-4S]3+ cluster with CoM-SH as a ligand. Subunit HdrB of the Methanothermobacter marburgensis HdrABC holoenzyme contains two cysteine-rich sequence motifs (CX31-39CCX35-36CXXC), designated as CCG domain in the Pfam database and conserved in many proteins. Here we present experimental evidence that the C-terminal CCG domain of HdrB binds this unusual [4Fe-4S] cluster. HdrB was produced in Escherichia coli, and an iron-sulfur cluster was subsequently inserted by in vitro reconstitution. In the oxidized state the cluster without the substrate exhibited a rhombic EPR signal (gzyx = 2.015, 1.995, and 1.950) reminiscent of the CoM-HDR signal. 57Fe ENDOR spectroscopy revealed that this paramagnetic species is a [4Fe-4S] cluster with 57Fe hyperfine couplings very similar to that of CoM-HDR. CoM-33SH resulted in a broadening of the EPR signal, and upon addition of CoM-SH the midpoint potential of the cluster was shifted to values observed for CoM-HDR, both indicating binding of CoM-SH to the cluster. Site-directed mutagenesis of all 12 cysteine residues in HdrB identified four cysteines of the C-terminal CCG domain as cluster ligands. Combined with the previous detection of CoM-HDR-like EPR signals in other CCG domain-containing proteins our data indicate a general role of the C-terminal CCG domain in coordination of this novel [4Fe-4S] cluster. In addition, Zn K-edge X-ray absorption spectroscopy identified an isolated Zn site with an S3(O/N)1 geometry in HdrB and the HDR holoenzyme. The N-terminal CCG domain is suggested to provide ligands to the Zn site.
Figures



Similar articles
-
Advanced electron paramagnetic resonance on the catalytic iron-sulfur cluster bound to the CCG domain of heterodisulfide reductase and succinate: quinone reductase.J Biol Inorg Chem. 2013 Dec;18(8):905-15. doi: 10.1007/s00775-013-1037-x. Epub 2013 Sep 14. J Biol Inorg Chem. 2013. PMID: 24037219
-
Coenzyme M binds to a [4Fe-4S] cluster in the active site of heterodisulfide reductase as deduced from EPR studies with the [33S]coenzyme M-treated enzyme.FEBS Lett. 2003 Mar 13;538(1-3):81-4. doi: 10.1016/s0014-5793(03)00134-0. FEBS Lett. 2003. PMID: 12633857
-
Heterodisulfide reductase from Methanothermobacter marburgensis contains an active-site [4Fe-4S] cluster that is directly involved in mediating heterodisulfide reduction.FEBS Lett. 2002 Feb 13;512(1-3):263-8. doi: 10.1016/s0014-5793(02)02281-0. FEBS Lett. 2002. PMID: 11852093
-
Heterodisulfide reductase from methanogenic archaea: a new catalytic role for an iron-sulfur cluster.Biol Chem. 2005 Oct;386(10):961-70. doi: 10.1515/BC.2005.112. Biol Chem. 2005. PMID: 16218868 Review.
-
Analysis of Mechanisms for Electron Uptake by Methanothrix harundinacea 6Ac During Direct Interspecies Electron Transfer.Int J Mol Sci. 2025 Apr 28;26(9):4195. doi: 10.3390/ijms26094195. Int J Mol Sci. 2025. PMID: 40362433 Free PMC article. Review.
Cited by
-
DsrMKJOP is the terminal reductase complex in anaerobic sulfate respiration.Proc Natl Acad Sci U S A. 2024 Feb 6;121(6):e2313650121. doi: 10.1073/pnas.2313650121. Epub 2024 Jan 29. Proc Natl Acad Sci U S A. 2024. PMID: 38285932 Free PMC article.
-
Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans.BMC Genomics. 2009 Aug 24;10:394. doi: 10.1186/1471-2164-10-394. BMC Genomics. 2009. PMID: 19703284 Free PMC article.
-
More than 200 genes required for methane formation from H₂ and CO₂ and energy conservation are present in Methanothermobacter marburgensis and Methanothermobacter thermautotrophicus.Archaea. 2011;2011:973848. doi: 10.1155/2011/973848. Epub 2011 Apr 27. Archaea. 2011. PMID: 21559116 Free PMC article.
-
Comparison of environmental and isolate Sulfobacillus genomes reveals diverse carbon, sulfur, nitrogen, and hydrogen metabolisms.BMC Genomics. 2014 Dec 15;15:1107. doi: 10.1186/1471-2164-15-1107. BMC Genomics. 2014. PMID: 25511286 Free PMC article.
-
Electron bifurcation involved in the energy metabolism of the acetogenic bacterium Moorella thermoacetica growing on glucose or H2 plus CO2.J Bacteriol. 2012 Jul;194(14):3689-99. doi: 10.1128/JB.00385-12. Epub 2012 May 11. J Bacteriol. 2012. PMID: 22582275 Free PMC article.
References
-
- Hedderich R, Klimmek O, Kröger A, Dirmeier R, Keller M, Stetter KO. Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol Rev. 1998;22:353–381.
-
- Thauer RK. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology. 1998;144:2377–2406. - PubMed
-
- Hedderich R, Berkessel A, Thauer RK. Purification and properties of heterodisulfide reductase from Methanobacterium thermoautotrophicum (strain Marburg) Eur J Biochem. 1990;193:255–261. - PubMed
-
- Künkel A, Vaupel M, Heim S, Thauer RK, Hedderich R. Heterodisulfide reductase from methanol-grown cells of Methanosarcina barkeri is not a flavoenzyme. Eur J Biochem. 1997;244:226–234. - PubMed
-
- Simianu M, Murakami E, Brewer JM, Ragsdale SW. Purification and properties of the heme- and iron-sulfur-containing heterodisulfide reductase from Methanosarcina thermophila. Biochemistry. 1998;37:10027–10039. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

