Rapid spread of mouse mammary tumor virus in cultured human breast cells
- PMID: 17931409
- PMCID: PMC2169256
- DOI: 10.1186/1742-4690-4-73
Rapid spread of mouse mammary tumor virus in cultured human breast cells
Abstract
Background: The role of mouse mammary tumor virus (MMTV) as a causative agent in human breast carcinogenesis has recently been the subject of renewed interest. The proposed model is based on the detection of MMTV sequences in human breast cancer but not in healthy breast tissue. One of the main drawbacks to this model, however, was that until now human cells had not been demonstrated to sustain productive MMTV infection.
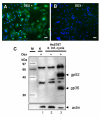
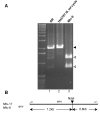
Results: Here, we show for the first time the rapid spread of mouse mammary tumor virus, MMTV(GR), in cultured human mammary cells (Hs578T), ultimately leading to the infection of every cell in culture. The replication of the virus was monitored by quantitative PCR, quantitative RT-PCR and immunofluorescence imaging. The infected human cells expressed, upon cultivation with dexamethasone, MMTV structural proteins and released spiked B-type virions, the infectivity of which could be neutralized by anti-MMTV antibody. Replication of the virus was efficiently blocked by an inhibitor of reverse transcription, 3'-azido-3'-deoxythymidine. The human origin of the infected cells was confirmed by determining a number of integration sites hosting the provirus, which were unequivocally identified as human sequences.
Conclusion: Taken together, our results show that human cells can support replication of mouse mammary tumor virus.
Figures







Similar articles
-
C3H strain of mouse mammary tumour virus, like GR strain, infects human mammary epithelial cells, albeit less efficiently than murine mammary epithelial cells.J Gen Virol. 2015 Mar;96(Pt 3):650-662. doi: 10.1099/jgv.0.000006. Epub 2014 Dec 9. J Gen Virol. 2015. PMID: 25491421
-
Presence of mouse mammary tumour-like virus gene sequences may be associated with morphology of specific human breast cancer.J Clin Pathol. 2006 Dec;59(12):1287-92. doi: 10.1136/jcp.2005.035907. Epub 2006 May 12. J Clin Pathol. 2006. PMID: 16698952 Free PMC article.
-
Reverse transcriptase-dependent and -independent phases of infection with mouse mammary tumor virus: implications for superantigen function.J Exp Med. 1994 Dec 1;180(6):2347-51. doi: 10.1084/jem.180.6.2347. J Exp Med. 1994. PMID: 7525852 Free PMC article.
-
A viral aetiology for breast cancer: time to re-examine the postulate.Intervirology. 2004;47(1):2-13. doi: 10.1159/000076636. Intervirology. 2004. PMID: 15044830 Review.
-
Of mice, cats, and men: is human breast cancer a zoonosis?Microsc Res Tech. 2005 Nov;68(3-4):197-208. doi: 10.1002/jemt.20232. Microsc Res Tech. 2005. PMID: 16276516 Review.
Cited by
-
The virology of breast cancer: viruses as the potential causative agents of breast tumorigenesis.Discov Med. 2019 Mar;27(148):163-166. Discov Med. 2019. PMID: 31095925 Free PMC article. Review.
-
Is MMTV associated with human breast cancer? Maybe, but probably not.Virol J. 2017 Oct 13;14(1):196. doi: 10.1186/s12985-017-0862-x. Virol J. 2017. PMID: 29029634 Free PMC article.
-
Common integration sites for MMTV in viral induced mouse mammary tumors.J Mammary Gland Biol Neoplasia. 2008 Sep;13(3):309-21. doi: 10.1007/s10911-008-9092-6. Epub 2008 Aug 15. J Mammary Gland Biol Neoplasia. 2008. PMID: 18709449 Free PMC article. Review.
-
Human RNA "rumor" viruses: the search for novel human retroviruses in chronic disease.Microbiol Mol Biol Rev. 2008 Mar;72(1):157-96, table of contents. doi: 10.1128/MMBR.00033-07. Microbiol Mol Biol Rev. 2008. PMID: 18322038 Free PMC article. Review.
-
Isolation of a Human Betaretrovirus from Patients with Primary Biliary Cholangitis.Viruses. 2022 Apr 24;14(5):886. doi: 10.3390/v14050886. Viruses. 2022. PMID: 35632628 Free PMC article.
References
-
- Sarkar NH. Type B virus and human breast cancer. In: Giraldo A and Beth A, editor. The role of viruses in human cancer. Vol. 1 , Elsevier, North Holland; 1980. pp. 207–235.
-
- Wang Y, Holland JF, Bleiweiss IJ, Melana S, Liu X, Pelisson I, Cantarella A, Stellrecht K, Mani S, Pogo BG. Detection of mammary tumor virus env gene-like sequences in human breast cancer. Cancer Res. 1995;55:5173–5179. - PubMed
-
- Etkind P, Du J, Khan A, Pillitteri J, Wiernik PH. Mouse mammary tumor virus-like ENV gene sequences in human breast tumors and in a lymphoma of a breast cancer patient. Clin Cancer Res. 2000;6:1273–1278. - PubMed
-
- Ford CE, Tran D, Deng Y, Ta VT, Rawlinson WD, Lawson JS. Mouse mammary tumor virus-like gene sequences in breast tumors of Australian and Vietnamese women. Clin Cancer Res. 2003;9:1118–1120. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

