Extent to which hairpin opening by the Artemis:DNA-PKcs complex can contribute to junctional diversity in V(D)J recombination
- PMID: 17932067
- PMCID: PMC2175297
- DOI: 10.1093/nar/gkm823
Extent to which hairpin opening by the Artemis:DNA-PKcs complex can contribute to junctional diversity in V(D)J recombination
Abstract
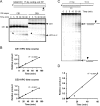
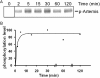
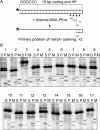
V(D)J recombination events are initiated by cleavage at gene segments by the RAG1:RAG2 complex, which results in hairpin formation at the coding ends. The hairpins are opened by the Artemis:DNA-PKcs complex, and then joined via the nonhomologous DNA end joining (NHEJ) process. Here we examine the opening of the hairpinned coding ends from all of the 39 functional human V(H) elements. We find that there is some sequence-dependent variation in the efficiency and even the position of hairpin opening by Artemis:DNA-PKcs. The hairpin opening efficiency varies over a 7-fold range. The hairpin opening position varies over the region from 1 to 4 nt 3' of the hairpin tip, leading to a 2-8 nt single-stranded 3' overhang at each coding end. This information provides greater clarity on the extent to which the hairpin opening position contributes to junctional diversification in V(D)J recombination.
Figures

References
-
- Swanson PC. The bounty of RAGs: recombination signal complexes and reaction outcomes. Immunol. Rev. 2004;200:90–114. - PubMed
-
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Ann. Rev. Immunol. 2000;18:495–527. - PubMed
-
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. - PubMed
-
- Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 2002;71:101–132. - PubMed
-
- Lieber MR. Site-specific recombination in the immune system. FASEB J. 1991;5:2934–2944. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

