Trans-lesion synthesis and RNaseH activity by reverse transcriptases on a true abasic RNA template
- PMID: 17932068
- PMCID: PMC2175328
- DOI: 10.1093/nar/gkm767
Trans-lesion synthesis and RNaseH activity by reverse transcriptases on a true abasic RNA template
Abstract
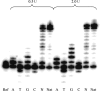
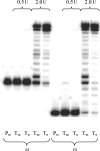
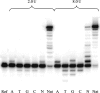
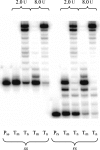
While much is known about abasic DNA, the biological impact of abasic RNA is largely unexplored. To test the mutagenic potential of this RNA lesion in the context of retroviruses, we synthesized a 31-mer oligoribonucleotide containing an abasic (rAS) site and used it as a template for studying DNA primer extension by HIV-1, avian myeloblastosis virus (AMV) and moloney murine leukemia virus (MMLV) reversed transcriptases (RT). We found that trans-lesion synthesis readily takes place with HIV-1 RT and to a lesser extent with AMV RT while MMLV RT aborts DNA synthesis. The preference of dNTP incorporation follows the order A approximately G > C approximately T and thus obeys to the 'A-rule'. In the case of HIV-1 RT, we measured the kinetic data of dNTP incorporation and compared it to abasic DNA. We found that A-incorporation is only 2-fold slower relative to a matched (undamaged) RNA template while it is 7-fold slower in the case of DNA. Furthermore, there is less discrimination in incorporation between the four dNTPs in the case of abasic RNA compared to abasic DNA. These experiments clearly point to a higher promiscuity of lesion bypass on abasic RNA. Given their known higher chemical stability, such rAS sites can clearly contribute to (retro)viral evolution.
Figures




Similar articles
-
Polymerization and RNase H activities of the reverse transcriptases from avian myeloblastosis, human immunodeficiency, and Moloney murine leukemia viruses are functionally uncoupled.J Biol Chem. 1991 Apr 25;266(12):7423-31. J Biol Chem. 1991. PMID: 1708386
-
Comparison of HIV-1 and avian myeloblastosis virus reverse transcriptase fidelity on RNA and DNA templates.J Biol Chem. 1992 May 25;267(15):10888-96. J Biol Chem. 1992. PMID: 1375233
-
Translesion synthesis by AMV, HIV, and MMLVreverse transcriptases using RNA templates containing inosine, guanosine, and their 8-oxo-7,8-dihydropurine derivatives.PLoS One. 2020 Aug 28;15(8):e0235102. doi: 10.1371/journal.pone.0235102. eCollection 2020. PLoS One. 2020. PMID: 32857764 Free PMC article.
-
[Inhibition of gene expression by antisense DNA].Nihon Rinsho. 1995 Mar;53(3):771-8. Nihon Rinsho. 1995. PMID: 7535367 Review. Japanese.
-
Nucleotide insertion and primer extension at abasic template sites in different sequence contexts.Ann N Y Acad Sci. 1994 Jul 29;726:132-42; discussion 142-3. doi: 10.1111/j.1749-6632.1994.tb52804.x. Ann N Y Acad Sci. 1994. PMID: 8092671 Review.
Cited by
-
Determinants of adenine-mutagenesis in diversity-generating retroelements.Nucleic Acids Res. 2021 Jan 25;49(2):1033-1045. doi: 10.1093/nar/gkaa1240. Nucleic Acids Res. 2021. PMID: 33367793 Free PMC article.
-
The effect of RNA base lesions on mRNA translation.Nucleic Acids Res. 2015 May 19;43(9):4713-20. doi: 10.1093/nar/gkv377. Epub 2015 Apr 20. Nucleic Acids Res. 2015. PMID: 25897124 Free PMC article.
-
RNA oxidation catalyzed by cytochrome c leads to its depurination and cross-linking, which may facilitate cytochrome c release from mitochondria.Free Radic Biol Med. 2012 Aug 15;53(4):854-62. doi: 10.1016/j.freeradbiomed.2012.05.044. Epub 2012 Jun 7. Free Radic Biol Med. 2012. PMID: 22683603 Free PMC article.
-
An assay for RNA oxidation induced abasic sites using the Aldehyde Reactive Probe.Free Radic Res. 2011 Feb;45(2):237-47. doi: 10.3109/10715762.2010.535529. Epub 2010 Nov 10. Free Radic Res. 2011. PMID: 21062214 Free PMC article.
-
An ancient influenza genome from Switzerland allows deeper insights into host adaptation during the 1918 flu pandemic in Europe.BMC Biol. 2025 Jul 1;23(1):179. doi: 10.1186/s12915-025-02282-z. BMC Biol. 2025. PMID: 40597331 Free PMC article.
References
-
- Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. - PubMed
-
- Lindahl T, Andersson A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 1972;11:3618–3623. - PubMed
-
- Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 1986;20:201–230. - PubMed
-
- Lhomme J, Constant JF, Demeunynck M. Abasic DNA structure, reactivity, and recognition. Biopolymers. 1999;52:65–83. - PubMed
-
- Schärer OD. Chemistry and biology of DNA repair. Angew. Chem. Int. Ed. 2003;42:2946–2974. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

