Mutation in the splicing factor Hprp3p linked to retinitis pigmentosa impairs interactions within the U4/U6 snRNP complex
- PMID: 17932117
- PMCID: PMC4494837
- DOI: 10.1093/hmg/ddm300
Mutation in the splicing factor Hprp3p linked to retinitis pigmentosa impairs interactions within the U4/U6 snRNP complex
Abstract
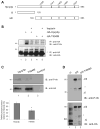
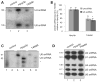
Mutations in PRPF3, a gene encoding the essential pre-mRNA splicing factor Hprp3p, have been identified in patients with autosomal dominant retinitis pigmentosa type 18 (RP18). Patients with RP18 have one of two single amino acid substitutions, Pro493Ser or Thr494Met, at the highly conserved Hprp3p C-terminal region. Pro493Ser occurs sporadically, whereas Thr494Met is observed in several unlinked RP families worldwide. The latter mutation also alters a potential recognition motif for phosphorylation by casein kinase II (CKII). To understand the molecular basis of RP18, we examined the consequences of Thr494Met mutation on Hprp3p molecular interactions with components of the U4/U6.U5 small nuclear ribonucleoprotein particles (snRNPs) complex. Since numerous mutations causing human diseases change pre-mRNA splice sites, we investigated whether Thr494Met substitution affects the processing of PRPF3 mRNA. We found that Thr494Met does not affect PRPF3 mRNA processing, indicating that the mutation may exert its effect primarily at the protein level. We used small hairpin RNAs to specifically silence the endogenous PRPF3 while simultaneously expressing HA-tagged Thr494Met. We demonstrated that the C- but not N-terminal region of Hprp3p is indeed phosphorylated by CKII in vitro and in cells. CKII-mediated Hprp3p phosphorylation was significantly reduced by Thr494Met mutation. Consequently, the Hprp3p C-terminal region is rendered partially defective in its association with itself, Hprp4p, and U4/U6 snRNA. Our findings provide new insights into the biology of Hprp3p and suggest that the loss of Hprp3p phosphorylation at Thr494 is a key step for initiating Thr494Met aberrant interactions within U4/U6 snRNP complex and that these are likely linked to the RP18 phenotype.
Conflict of interest statement
Figures
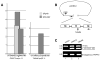





References
-
- Wang Q, Chen Q, Zhao K, Wang L, Traboulsi EI. Update on the molecular genetics of retinitis pigmentosa. Ophthalmic Genet. 2001;22:133–154. - PubMed
-
- Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. - PubMed
-
- McKie AB, McHale JC, Keen TJ, Tarttelin EE, Goliath R, van Lith-Verhoeven JJ, Greenberg J, Ramesar RS, Hoyng CB, Cremers FP, et al. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13) Hum Mol Genet. 2001;10:1555–1562. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

