Electrostatic rate enhancement and transient complex of protein-protein association
- PMID: 17932929
- PMCID: PMC3526769
- DOI: 10.1002/prot.21679
Electrostatic rate enhancement and transient complex of protein-protein association
Abstract
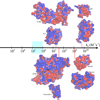
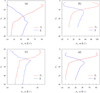
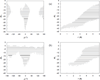
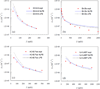
The association of two proteins is bounded by the rate at which they, via diffusion, find each other while in appropriate relative orientations. Orientational constraints restrict this rate to approximately 10(5)-10(6) M(-1) s(-1). Proteins with higher association rates generally have complementary electrostatic surfaces; proteins with lower association rates generally are slowed down by conformational changes upon complex formation. Previous studies (Zhou, Biophys J 1997;73:2441-2445) have shown that electrostatic enhancement of the diffusion-limited association rate can be accurately modeled by $k_{\bf D}$ = $k_{D}0\ {exp} ( - \langle U_{el} \rangle;{\star}/k_{B} T),$ where k(D) and k(D0) are the rates in the presence and absence of electrostatic interactions, respectively, U(el) is the average electrostatic interaction energy in a "transient-complex" ensemble, and k(B)T is the thermal energy. The transient-complex ensemble separates the bound state from the unbound state. Predictions of the transient-complex theory on four protein complexes were found to agree well with the experiment when the electrostatic interaction energy was calculated with the linearized Poisson-Boltzmann (PB) equation (Alsallaq and Zhou, Structure 2007;15:215-224). Here we show that the agreement is further improved when the nonlinear PB equation is used. These predictions are obtained with the dielectric boundary defined as the protein van der Waals surface. When the dielectric boundary is instead specified as the molecular surface, electrostatic interactions in the transient complex become repulsive and are thus predicted to retard association. Together these results demonstrate that the transient-complex theory is predictive of electrostatic rate enhancement and can help parameterize PB calculations.
(c) 2007 Wiley-Liss, Inc.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

