ProSAS: a database for analyzing alternative splicing in the context of protein structures
- PMID: 17933774
- PMCID: PMC2238869
- DOI: 10.1093/nar/gkm793
ProSAS: a database for analyzing alternative splicing in the context of protein structures
Abstract
Alternative splicing is known to be one of the major sources for functional diversity in higher eukaryotes. Several splicing isoforms have been characterized in the literature that play important roles in cellular processes like apoptosis or signal transduction pathways. Splicing events can often be detected on the mRNA level by large-scale cDNA or EST experiments and such data is collected and annotated in several databases. Nevertheless, the effects of splicing on the structure of a protein are largely unknown. The ProSAS (Protein Structure and Alternative Splicing) database fills this gap and provides a unified resource for analyzing effects of alternative splicing events in the context of protein structures. ProSAS comprehensively annotates and models protein structures for several Ensembl genomes as well as SwissProt entries harbouring splicing events. Alternative isoforms annotated in Ensembl or SwissProt can be analyzed on the protein structure and protein function level using an intuitive user interface that provides several features and tools for a structure-based analysis of alternative splicing events. The ProSAS database is freely accessible at http://www.bio.ifi.lmu.de/ProSAS.
Figures
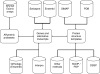
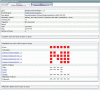
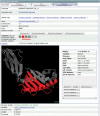
Similar articles
-
AS-ALPS: a database for analyzing the effects of alternative splicing on protein structure, interaction and network in human and mouse.Nucleic Acids Res. 2009 Jan;37(Database issue):D305-9. doi: 10.1093/nar/gkn869. Epub 2008 Nov 10. Nucleic Acids Res. 2009. PMID: 19015123 Free PMC article.
-
APPRIS: annotation of principal and alternative splice isoforms.Nucleic Acids Res. 2013 Jan;41(Database issue):D110-7. doi: 10.1093/nar/gks1058. Epub 2012 Nov 17. Nucleic Acids Res. 2013. PMID: 23161672 Free PMC article.
-
Alternative splicing and protein structure evolution.Nucleic Acids Res. 2008 Feb;36(2):550-8. doi: 10.1093/nar/gkm1054. Epub 2007 Nov 30. Nucleic Acids Res. 2008. PMID: 18055499 Free PMC article.
-
Increase of functional diversity by alternative splicing.Trends Genet. 2003 Mar;19(3):124-8. doi: 10.1016/S0168-9525(03)00023-4. Trends Genet. 2003. PMID: 12615003 Review.
-
Exploring the functional impact of alternative splicing on human protein isoforms using available annotation sources.Brief Bioinform. 2019 Sep 27;20(5):1754-1768. doi: 10.1093/bib/bby047. Brief Bioinform. 2019. PMID: 29931155 Free PMC article. Review.
Cited by
-
AS-ALPS: a database for analyzing the effects of alternative splicing on protein structure, interaction and network in human and mouse.Nucleic Acids Res. 2009 Jan;37(Database issue):D305-9. doi: 10.1093/nar/gkn869. Epub 2008 Nov 10. Nucleic Acids Res. 2009. PMID: 19015123 Free PMC article.
-
APPRIS: annotation of principal and alternative splice isoforms.Nucleic Acids Res. 2013 Jan;41(Database issue):D110-7. doi: 10.1093/nar/gks1058. Epub 2012 Nov 17. Nucleic Acids Res. 2013. PMID: 23161672 Free PMC article.
-
Genomic landscape of developing male germ cells.Birth Defects Res C Embryo Today. 2009 Mar;87(1):43-63. doi: 10.1002/bdrc.20147. Birth Defects Res C Embryo Today. 2009. PMID: 19306351 Free PMC article. Review.
-
Alternative splicing and protein structure evolution.Nucleic Acids Res. 2008 Feb;36(2):550-8. doi: 10.1093/nar/gkm1054. Epub 2007 Nov 30. Nucleic Acids Res. 2008. PMID: 18055499 Free PMC article.
-
ExDom: an integrated database for comparative analysis of the exon-intron structures of protein domains in eukaryotes.Nucleic Acids Res. 2009 Jan;37(Database issue):D703-11. doi: 10.1093/nar/gkn746. Epub 2008 Nov 4. Nucleic Acids Res. 2009. PMID: 18984624 Free PMC article.
References
-
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

