Subcellular localization and functional domain studies of DEFECTIVE KERNEL1 in maize and Arabidopsis suggest a model for aleurone cell fate specification involving CRINKLY4 and SUPERNUMERARY ALEURONE LAYER1
- PMID: 17933905
- PMCID: PMC2174714
- DOI: 10.1105/tpc.106.048868
Subcellular localization and functional domain studies of DEFECTIVE KERNEL1 in maize and Arabidopsis suggest a model for aleurone cell fate specification involving CRINKLY4 and SUPERNUMERARY ALEURONE LAYER1
Abstract
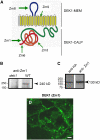
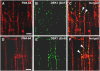
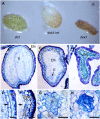
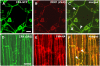
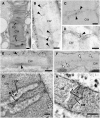
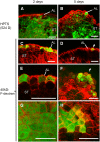
DEFECTIVE KERNEL1 (DEK1), which consists of a membrane-spanning region (DEK1-MEM) and a calpain-like Cys proteinase region (DEK1-CALP), is essential for aleurone cell formation at the surface of maize (Zea mays) endosperm. Immunolocalization and FM4-64 dye incubation experiments showed that DEK1 and CRINKLY4 (CR4), a receptor kinase implicated in aleurone cell fate specification, colocalized to plasma membrane and endosomes. SUPERNUMERARY ALEURONE LAYER1 (SAL1), a negative regulator of aleurone cell fate encoding a class E vacuolar sorting protein, colocalized with DEK1 and CR4 in endosomes. Immunogold localization, dual-axis electron tomography, and diffusion of fluorescent dye tracers showed that young aleurone cells established symplastic subdomains through plasmodesmata of larger dimensions than those connecting starchy endosperm cells and that CR4 preferentially associated with plasmodesmata between aleurone cells. Genetic complementation experiments showed that DEK1-CALP failed to restore wild-type phenotypes in maize and Arabidopsis thaliana dek1 mutants, and DEK1-MEM also failed to restore wild-type phenotypes in Arabidopsis dek1-1 mutants. Instead, ectopic expression of DEK1-MEM under the control of the cauliflower mosaic virus 35S promoter gave a dominant negative phenotype. These data suggest a model for aleurone cell fate specification in which DEK1 perceives and/or transmits a positional signal, CR4 promotes the lateral movement of aleurone signaling molecules between aleurone cells, and SAL1 maintains the proper plasma membrane concentration of DEK1 and CR4 proteins via endosome-mediated recycling/degradation.
Figures





Similar articles
-
The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily.Proc Natl Acad Sci U S A. 2002 Apr 16;99(8):5460-5. doi: 10.1073/pnas.042098799. Epub 2002 Apr 2. Proc Natl Acad Sci U S A. 2002. PMID: 11929961 Free PMC article.
-
The maize dek1 gene functions in embryonic pattern formation and cell fate specification.Development. 2002 Nov;129(22):5217-25. doi: 10.1242/dev.129.22.5217. Development. 2002. PMID: 12399313
-
The thick aleurone1 mutant defines a negative regulation of maize aleurone cell fate that functions downstream of defective kernel1.Plant Physiol. 2011 Aug;156(4):1826-36. doi: 10.1104/pp.111.177725. Epub 2011 May 26. Plant Physiol. 2011. PMID: 21617032 Free PMC article.
-
Sensing and transducing forces in plants with MSL10 and DEK1 mechanosensors.FEBS Lett. 2018 Jun;592(12):1968-1979. doi: 10.1002/1873-3468.13102. Epub 2018 Jun 4. FEBS Lett. 2018. PMID: 29782638 Review.
-
Differentiation mechanism and function of the cereal aleurone cells and hormone effects on them.Plant Cell Rep. 2014 Nov;33(11):1779-87. doi: 10.1007/s00299-014-1654-z. Epub 2014 Jul 10. Plant Cell Rep. 2014. PMID: 25007781 Review.
Cited by
-
Artificial ubiquitylation is sufficient for sorting of a plasma membrane ATPase to the vacuolar lumen of Arabidopsis cells.Planta. 2012 Jul;236(1):63-77. doi: 10.1007/s00425-012-1587-0. Epub 2012 Jan 19. Planta. 2012. PMID: 22258747
-
A phylogenetic approach to study the origin and evolution of the CRINKLY4 family.Front Plant Sci. 2015 Oct 23;6:880. doi: 10.3389/fpls.2015.00880. eCollection 2015. Front Plant Sci. 2015. PMID: 26557128 Free PMC article.
-
Massive expansion of the calpain gene family in unicellular eukaryotes.BMC Evol Biol. 2012 Sep 29;12:193. doi: 10.1186/1471-2148-12-193. BMC Evol Biol. 2012. PMID: 23020305 Free PMC article.
-
Suppressor of K+ transport growth defect 1 (SKD1) interacts with RING-type ubiquitin ligase and sucrose non-fermenting 1-related protein kinase (SnRK1) in the halophyte ice plant.J Exp Bot. 2013 May;64(8):2385-400. doi: 10.1093/jxb/ert097. Epub 2013 Apr 11. J Exp Bot. 2013. PMID: 23580756 Free PMC article.
-
CLE peptides act via the receptor-like kinase CRINKLY 4 in Physcomitrium patens gametophore development.Plant Signal Behav. 2024 Dec 31;19(1):2386502. doi: 10.1080/15592324.2024.2386502. Epub 2024 Jul 31. Plant Signal Behav. 2024. PMID: 39082799 Free PMC article.
References
-
- Aggarwal, B.B. (2003). Signalling pathways of the TNF superfamily: A double-edged sword. Nat. Rev. Immunol. 3 745–756. - PubMed
-
- Babst, M. (2005). A protein's final ESCRT. Traffic 6 2–9. - PubMed
-
- Becraft, P.W., and Asuncion-Crabb, Y. (2000). Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development 127 4039–4048. - PubMed
-
- Becraft, P.W., Li, K., Dey, N., and Asuncion-Crabb, Y. (2002). The maize Dek1 gene functions in embryonic pattern formation and cell fate specification. Development 129 5217–5225. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

