Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II
- PMID: 17933913
- PMCID: PMC2168159
- DOI: 10.1128/AEM.01428-07
Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II
Abstract
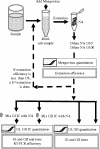
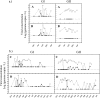
Noroviruses, an important cause of gastroenteritis, are excreted by infected individuals and are therefore present in wastewater. We quantified norovirus genogroup I (GI) and GII in wastewater at different locations in France and evaluated removal by a range of treatment types, including basic (waste stabilization pond), current industry standard (activated sludge), and state-of-the-art (submerged membrane bioreactor) treatments. Noroviruses were quantified using real-time reverse transcription-PCR (rRT-PCR). Mengovirus was used as a virus extraction control, and internal controls were used to verify the level of GI and GII rRT-PCR inhibition. A total of 161 (81 influent and 79 effluent) samples were examined; GI and GII were detected in 43 and 88% of the influent samples, respectively, and in 24 and 14% of the effluent samples, respectively. Physicians in France report far more cases of GII than GI during outbreaks; thus, the frequent presence of GI was unexpected. The GI influent concentrations were more variable, the peak GI influent concentrations were higher than the peak GII influent concentrations at all four sites (up to 1 x 10(9) and 6 x 10(7) genome copies/liter, respectively), and the average positive influent concentrations of GI were higher than the average positive influent concentrations of GII. The maximum effluent breakthrough concentrations were 6 x 10(6) and 3 x 10(6) genome copies/liter for GI and GII, respectively, indicating that the four treatment systems studied decreased the norovirus contamination load in receiving waters.
Figures
References
-
- Atmar, R. L., and M. K. Estes. 2006. The epidemiologic and clinical importance of norovirus infection. Gastroenterol. Clin. N. Am. 35:275-290. - PubMed
-
- Blanton, L. H., S. M. Adams, R. S. Beard, G. Wei, S. N. Bulens, M. A. Widdowson, R. I. Glass, and S. S. Monroe. 2006. Molecular and epidemiologic trends of caliciviruses associated with outbreaks of acute gastroenteritis in the United States, 2000-2004. J. Infect. Dis. 193:413-421. - PubMed
-
- Bon, F., K. Ambert-Balay, H. Giraudon, J. Kaplon, S. Le Guyader, M. Pommepuy, A. Gallay, V. Vaillant, H. de Valk, R. Chikhi-Brachet, A. Flahaut, P. Pothier, and E. Kohli. 2005. Molecular epidemiology of caliciviruses detected in sporadic and outbreak cases of gastroenteritis in France from December 1998 to February 2004. J. Clin. Microbiol. 43:4659-4664. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

