Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1
- PMID: 17933933
- PMCID: PMC2168065
- DOI: 10.1128/AEM.01649-07
Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1
Abstract
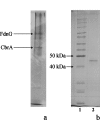
A chlorobenzene reductive dehalogenase of the anaerobic dehalorespiring bacterium Dehalococcoides sp. strain CBDB1 was identified. Due to poor biomass yields, standard protein isolation procedures were not applicable. Therefore, cell extracts from cultures grown on trichlorobenzenes were separated by native polyacrylamide gel electrophoresis and analyzed directly for chlorobenzene reductive dehalogenase activity within gel fragments. Activity was found in a single band, even though electrophoretic separation was performed under aerobic conditions. Matrix-assisted laser desorption ionization mass spectrometry (MALDI MS) and nano-liquid chromatography-MALDI MS analysis of silver-stained replicas of the active band on native polyacrylamide gels identified a protein product of the cbdbA84 gene, now called cbrA. The cbdbA84 gene is one of 32 reductive dehalogenase homologous genes present in the genome of strain CBDB1. The chlorobenzene reductive dehalogenase identified in our study represents a member of the family of corrinoid/iron-sulfur cluster-containing reductive dehalogenases. No orthologs of cbdbA84 were found in the completely sequenced genomes of Dehalococcoides sp. strains 195 and BAV1 nor among the genes amplified from Dehalococcoides sp. strain FL2 or mixed cultures containing Dehalococcoides. Another dehalogenase homologue (cbdbA80) was expressed in cultures that contained 1,2,4-trichlorobenzene, but its role is unclear. Other highly expressed proteins identified with our approach included the major subunit of a protein annotated as formate dehydrogenase, transporter subunits, and a putative S-layer protein.
Figures

Similar articles
-
Multiple nonidentical reductive-dehalogenase-homologous genes are common in Dehalococcoides.Appl Environ Microbiol. 2004 Sep;70(9):5290-7. doi: 10.1128/AEM.70.9.5290-5297.2004. Appl Environ Microbiol. 2004. PMID: 15345412 Free PMC article.
-
Dehalogenimonas sp. Strain WBC-2 Genome and Identification of Its trans-Dichloroethene Reductive Dehalogenase, TdrA.Appl Environ Microbiol. 2015 Oct 9;82(1):40-50. doi: 10.1128/AEM.02017-15. Print 2016 Jan 1. Appl Environ Microbiol. 2015. PMID: 26452554 Free PMC article.
-
A H2 -oxidizing, 1,2,3-trichlorobenzene-reducing multienzyme complex isolated from the obligately organohalide-respiring bacterium Dehalococcoides mccartyi strain CBDB1.Environ Microbiol Rep. 2017 Oct;9(5):618-625. doi: 10.1111/1758-2229.12560. Epub 2017 Jul 13. Environ Microbiol Rep. 2017. PMID: 28631290
-
Biochemical and genetic bases of dehalorespiration.Chem Rec. 2008;8(1):1-12. doi: 10.1002/tcr.20134. Chem Rec. 2008. PMID: 18302277 Review.
-
Reductive Dehalogenases Come of Age in Biological Destruction of Organohalides.Trends Biotechnol. 2015 Oct;33(10):595-610. doi: 10.1016/j.tibtech.2015.07.004. Trends Biotechnol. 2015. PMID: 26409778 Review.
Cited by
-
Genome sequence of the organohalide-respiring Dehalogenimonas alkenigignens type strain (IP3-3(T)).Stand Genomic Sci. 2016 Jun 23;11:44. doi: 10.1186/s40793-016-0165-7. eCollection 2016. Stand Genomic Sci. 2016. PMID: 27340512 Free PMC article.
-
Hexachlorobenzene Monooxygenase Substrate Selectivity and Catalysis: Structural and Biochemical Insights.Appl Environ Microbiol. 2020 Dec 17;87(1):e01965-20. doi: 10.1128/AEM.01965-20. Print 2020 Dec 17. Appl Environ Microbiol. 2020. PMID: 33097503 Free PMC article.
-
Changes of the Proteome and Acetylome during Transition into the Stationary Phase in the Organohalide-Respiring Dehalococcoides mccartyi Strain CBDB1.Microorganisms. 2021 Feb 12;9(2):365. doi: 10.3390/microorganisms9020365. Microorganisms. 2021. PMID: 33673241 Free PMC article.
-
Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides.PLoS Genet. 2009 Nov;5(11):e1000714. doi: 10.1371/journal.pgen.1000714. Epub 2009 Nov 6. PLoS Genet. 2009. PMID: 19893622 Free PMC article.
-
Mechanisms of aerobic dechlorination of hexachlorobenzene and pentachlorophenol by Nocardioides sp. PD653.J Pestic Sci. 2021 Nov 20;46(4):373-381. doi: 10.1584/jpestics.J21-04. J Pestic Sci. 2021. PMID: 34908898 Free PMC article.
References
-
- Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. - PubMed
-
- Adrian, L., S. K. Hansen, J. M. Fung, H. Görisch, and S. H. Zinder. 2007. Growth of Dehalococcoides strains with chlorophenols as electron acceptor. Environ. Sci. Technol. 41:2318-2323. - PubMed
-
- Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. - PubMed
-
- Bunge, M., L. Adrian, A. Kraus, M. Opel, W. G. Lorenz, J. R. Andreesen, H. Görisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357-360. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

