Thermodynamic analysis shows conformational coupling and dynamics confer substrate specificity in fructose-1,6-bisphosphate aldolase
- PMID: 17935305
- PMCID: PMC2546497
- DOI: 10.1021/bi700713s
Thermodynamic analysis shows conformational coupling and dynamics confer substrate specificity in fructose-1,6-bisphosphate aldolase
Abstract
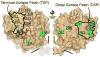
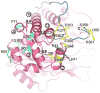
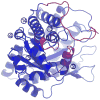
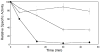
Conformational flexibility is emerging as a central theme in enzyme catalysis. Thus, identifying and characterizing enzyme dynamics are critical for understanding catalytic mechanisms. Herein, coupling analysis, which uses thermodynamic analysis to assess cooperativity and coupling between distal regions on an enzyme, is used to interrogate substrate specificity among fructose-1,6-(bis)phosphate aldolase (aldolase) isozymes. Aldolase exists as three isozymes, A, B, and C, distinguished by their unique substrate preferences despite the fact that the structures of the active sites of the three isozymes are nearly identical. While conformational flexibility has been observed in aldolase A, its function in the catalytic reaction of aldolase has not been demonstrated. To explore the role of conformational dynamics in substrate specificity, those residues associated with isozyme specificity (ISRs) were swapped and the resulting chimeras were subjected to steady-state kinetics. Thermodynamic analyses suggest cooperativity between a terminal surface patch (TSP) and a distal surface patch (DSP) of ISRs that are separated by >8.9 A. Notably, the coupling energy (DeltaGI) is anticorrelated with respect to the two substrates, fructose 1,6-bisphosphate and fructose 1-phosphate. The difference in coupling energy with respect to these two substrates accounts for approximately 70% of the energy difference for the ratio of kcat/Km for the two substrates between aldolase A and aldolase B. These nonadditive mutational effects between the TSP and DSP provide functional evidence that coupling interactions arising from conformational flexibility during catalysis are a major determinant of substrate specificity.
Figures


Similar articles
-
Spatial clustering of isozyme-specific residues reveals unlikely determinants of isozyme specificity in fructose-1,6-bisphosphate aldolase.J Biol Chem. 2003 May 9;278(19):17307-13. doi: 10.1074/jbc.M209185200. Epub 2003 Feb 28. J Biol Chem. 2003. PMID: 12611890
-
Exploring substrate binding and discrimination in fructose1, 6-bisphosphate and tagatose 1,6-bisphosphate aldolases.Eur J Biochem. 2000 Mar;267(6):1858-68. doi: 10.1046/j.1432-1327.2000.01191.x. Eur J Biochem. 2000. PMID: 10712619
-
Structure of human brain fructose 1,6-(bis)phosphate aldolase: linking isozyme structure with function.Protein Sci. 2004 Dec;13(12):3077-84. doi: 10.1110/ps.04915904. Epub 2004 Nov 10. Protein Sci. 2004. PMID: 15537755 Free PMC article.
-
Structure of a fructose-1,6-bis(phosphate) aldolase liganded to its natural substrate in a cleavage-defective mutant at 2.3 A(,).Biochemistry. 1999 Sep 28;38(39):12655-64. doi: 10.1021/bi9828371. Biochemistry. 1999. PMID: 10504235
-
Sweet siblings with different faces: the mechanisms of FBP and F6P aldolase, transaldolase, transketolase and phosphoketolase revisited in light of recent structural data.Bioorg Chem. 2014 Dec;57:263-280. doi: 10.1016/j.bioorg.2014.09.001. Epub 2014 Sep 16. Bioorg Chem. 2014. PMID: 25267444 Review.
Cited by
-
Uptake and metabolism of fructose by rat neocortical cells in vivo and by isolated nerve terminals in vitro.J Neurochem. 2015 May;133(4):572-81. doi: 10.1111/jnc.13079. Epub 2015 Mar 13. J Neurochem. 2015. PMID: 25708447 Free PMC article.
-
Aldolase directly interacts with ARNO and modulates cell morphology and acidic vesicle distribution.Am J Physiol Cell Physiol. 2011 Jun;300(6):C1442-55. doi: 10.1152/ajpcell.00076.2010. Epub 2011 Feb 9. Am J Physiol Cell Physiol. 2011. PMID: 21307348 Free PMC article.
-
Detection of native-state nonadditivity in double mutant cycles via hydrogen exchange.J Am Chem Soc. 2010 Jun 16;132(23):8010-9. doi: 10.1021/ja1003922. J Am Chem Soc. 2010. PMID: 20481530 Free PMC article.
-
The Network Basis for the Structural Thermostability and the Functional Thermoactivity of Aldolase B.Molecules. 2023 Feb 15;28(4):1850. doi: 10.3390/molecules28041850. Molecules. 2023. PMID: 36838836 Free PMC article.
-
Monitoring aromatic picosecond to nanosecond dynamics in proteins via 13C relaxation: expanding perturbation mapping of the rigidifying core mutation, V54A, in eglin c.Biochemistry. 2008 Apr 29;47(17):4876-86. doi: 10.1021/bi702330t. Epub 2008 Apr 5. Biochemistry. 2008. PMID: 18393447 Free PMC article.
References
-
- Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–541. - PubMed
-
- Schnell JR, Dyson HJ, Wright PE. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu Rev Biophys Biomol Struct. 2004;33:119–140. - PubMed
-
- Wand AJ. Dynamic activation of protein function: a view emerging from NMR spectroscopy. Nat Struct Biol. 2001;8:926–931. - PubMed
-
- Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. - PubMed
-
- Cooper A, Dryden DT. Allostery without conformational change. A plausible model. Eur Biophys J. 1984;11:103–109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

