Design of DNA minor groove binding diamidines that recognize GC base pair sequences: a dimeric-hinge interaction motif
- PMID: 17935330
- PMCID: PMC3865524
- DOI: 10.1021/ja074560a
Design of DNA minor groove binding diamidines that recognize GC base pair sequences: a dimeric-hinge interaction motif
Abstract
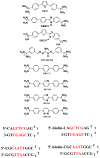
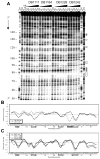
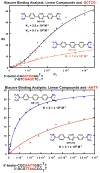
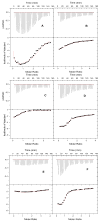
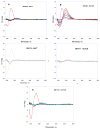
The classical model of DNA minor groove binding compounds is that they should have a crescent shape that closely fits the helical twist of the groove. Several compounds with relatively linear shape and large dihedral twist, however, have been found recently to bind strongly to the minor groove. These observations raise the question of how far the curvature requirement could be relaxed. As an initial step in experimental analysis of this question, a linear triphenyl diamidine, DB1111, and a series of nitrogen tricyclic analogues were prepared. The goal with the heterocycles is to design GC binding selectivity into heterocyclic compounds that can get into cells and exert biological effects. The compounds have a zero radius of curvature from amidine carbon to amidine carbon but a significant dihedral twist across the tricyclic and amidine-ring junctions. They would not be expected to bind well to the DNA minor groove by shape-matching criteria. Detailed DNase I footprinting studies of the sequence specificity of this set of diamidines indicated that a pyrimidine heterocyclic derivative, DB1242, binds specifically to a GC-rich sequence, -GCTCG-. It binds to the GC sequence more strongly than to the usual AT recognition sequences for curved minor groove agents. Other similar derivatives did not exhibit the GC specificity. Biosensor-surface plasmon resonance and isothermal titration calorimetry experiments indicate that DB1242 binds to the GC sequence as a highly cooperative stacked dimer. Circular dichroism results indicate that the compound binds in the minor groove. Molecular modeling studies support a minor groove complex and provide an inter-compound and compound-DNA hydrogen-bonding rational for the unusual GC binding specificity and the requirement for a pyrimidine heterocycle. This compound represents a new direction in the development of DNA sequence-specific agents, and it is the first non-polyamide, synthetic compound to specifically recognize a DNA sequence with a majority of GC base pairs.
Figures

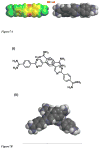
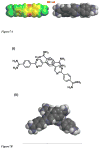
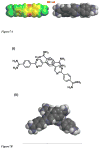
Similar articles
-
Heterocyclic diamidine interactions at AT base pairs in the DNA minor groove: effects of heterocycle differences, DNA AT sequence and length.J Phys Chem B. 2008 Sep 18;112(37):11809-18. doi: 10.1021/jp804048c. Epub 2008 Aug 22. J Phys Chem B. 2008. PMID: 18717551 Free PMC article.
-
Induced fit conformational changes of a "reversed amidine" heterocycle: optimized interactions in a DNA minor groove complex.J Am Chem Soc. 2007 May 2;129(17):5688-98. doi: 10.1021/ja069003n. Epub 2007 Apr 11. J Am Chem Soc. 2007. PMID: 17425312 Free PMC article.
-
Evaluation of the influence of compound structure on stacked-dimer formation in the DNA minor groove.Biochemistry. 2001 Feb 27;40(8):2511-21. doi: 10.1021/bi002301r. Biochemistry. 2001. PMID: 11327873
-
Binding to the DNA Minor Groove by Heterocyclic Dications: from AT Specific to GC Recognition Compounds.Curr Protoc. 2023 Apr;3(4):e729. doi: 10.1002/cpz1.729. Curr Protoc. 2023. PMID: 37071034 Review.
-
Binding to the DNA minor groove by heterocyclic dications: from AT-specific monomers to GC recognition with dimers.Curr Protoc Nucleic Acid Chem. 2012 Dec;Chapter 8:Unit8.8. doi: 10.1002/0471142700.nc0808s51. Curr Protoc Nucleic Acid Chem. 2012. PMID: 23255206 Free PMC article. Review.
Cited by
-
Using Genome Sequence to Enable the Design of Medicines and Chemical Probes.Chem Rev. 2018 Feb 28;118(4):1599-1663. doi: 10.1021/acs.chemrev.7b00504. Epub 2018 Jan 11. Chem Rev. 2018. PMID: 29322778 Free PMC article. Review.
-
Sensitive and Selective Detection of HIV-1 RRE RNA Using Vertical Silicon Nanowire Electrode Array.Nanoscale Res Lett. 2016 Dec;11(1):341. doi: 10.1186/s11671-016-1504-8. Epub 2016 Jul 22. Nanoscale Res Lett. 2016. PMID: 27448026 Free PMC article.
-
Influence of DNA structure on adjacent site cooperative binding.J Phys Chem B. 2008 Jul 24;112(29):8770-8. doi: 10.1021/jp801997v. Epub 2008 Jun 27. J Phys Chem B. 2008. PMID: 18582108 Free PMC article.
-
Protein Arginine Methylation and Citrullination in Epigenetic Regulation.ACS Chem Biol. 2016 Mar 18;11(3):654-68. doi: 10.1021/acschembio.5b00942. Epub 2015 Dec 31. ACS Chem Biol. 2016. PMID: 26686581 Free PMC article.
-
A role for water molecules in DNA-ligand minor groove recognition.Acc Chem Res. 2009 Jan 20;42(1):11-21. doi: 10.1021/ar800016q. Acc Chem Res. 2009. PMID: 18798655 Free PMC article.
References
-
- Russ AP, Lampel S. Drug Discov Today. 2005;10:1607–10. - PubMed
-
- Hajduk PJ, Huth JR, Tse C. Drug Discov Today. 2005;10:1675–82. - PubMed
-
- Lansiaux A, Tanious F, Mishal Z, Dassonneville L, Kumar A, Stephens CE, Hu Q, Wilson WD, Boykin DW, Bailly C. Cancer Res. 2002;62:7219–29. - PubMed
-
- Lansiaux A, Dassonneville L, Facompre M, Kumar A, Stephens CE, Bajic M, Tanious F, Wilson WD, Boykin DW, Bailly C. J Med Chem. 2002;45:1994–2002. - PubMed
-
- Susbielle G, Blattes R, Brevet V, Monod C, Kas E. Curr Med Chem Anticancer Agents. 2005;5:409–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

