DNA indels in coding regions reveal selective constraints on protein evolution in the human lineage
- PMID: 17935613
- PMCID: PMC2151769
- DOI: 10.1186/1471-2148-7-191
DNA indels in coding regions reveal selective constraints on protein evolution in the human lineage
Abstract
Background: Insertions and deletions of DNA segments (indels) are together with substitutions the major mutational processes that generate genetic variation. Here we focus on recent DNA insertions and deletions in protein coding regions of the human genome to investigate selective constraints on indels in protein evolution.
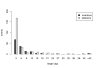
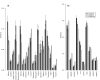
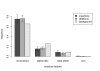
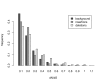
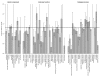
Results: Frequencies of inserted and deleted amino acids differ from background amino acid frequencies in the human proteome. Small amino acids are overrepresented, while hydrophobic, aliphatic and aromatic amino acids are strongly suppressed. Indels are found to be preferentially located in protein regions that do not form important structural domains. Amino acid insertion and deletion rates in genes associated with elementary biochemical reactions (e. g. catalytic activity, ligase activity, electron transport, or catabolic process) are lower compared to those in other genes and are therefore subject to stronger purifying selection.
Conclusion: Our analysis indicates that indels in human protein coding regions are subject to distinct levels of selective pressure with regard to their structural impact on the amino acid sequence, as well as to general properties of the genes they are located in. These findings confirm that many commonly accepted characteristics of selective constraints for substitutions are also valid for amino acid insertions and deletions.
Figures
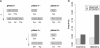
References
-
- Table of non-frameshifting indels in protein coding regions of the human genome. http://evogen.molgen.mpg.de/data/coding_indels41.txt
-
- Chen FC, Li WH. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet. 2001;7(2):444–456. doi: 10.1086/318206. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pub%med&pubmedi... - DOI - PMC - PubMed
-
- Britten RJ, Rowen L, Williams J, Cameron RA. Majority of divergence between closely related DNA samples is due to indels. Proc Natl Acad Sci USA. 2003;7(8):4661–4665. doi: 10.1073/pnas.0330964100. http://dx.doi.org/10.1073/pnas.0330964100 - DOI - DOI - PMC - PubMed
-
- Watanabe H, Fujiyama A, Hattori M, Taylor TD, Toyoda A, Kuroki Y, Noguchi H, BenKahla A, Lehrach H, Sudbrak R, Kube M, Taenzer S, Galgoczy P, Platzer M, Scharfe M, Nordsiek G, Blöcker H, Hellmann I, Khaitovich P, Pääbo S, Reinhardt R, Zheng HJ, Zhang XL, Zhu GF, Wang BF, Fu G, Ren SX, Zhao GP, Chen Z, Lee YS, Cheong JE, Choi SH, Wu KM, Liu TT, Hsiao KJ, Tsai SF, Kim CG, OOta S, Kitano T, Kohara Y, Saitou N, Park HS, Wang SY, Yaspo ML, Sakaki Y. DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature. 2004;7(6990):382–388. doi: 10.1038/nature02564. http://dx.doi.org/10.1038/nature02564 - DOI - DOI - PubMed
LinkOut - more resources
Full Text Sources

