Transcriptional networks in plasmacytoid dendritic cells stimulated with synthetic TLR 7 agonists
- PMID: 17935622
- PMCID: PMC2175514
- DOI: 10.1186/1471-2172-8-26
Transcriptional networks in plasmacytoid dendritic cells stimulated with synthetic TLR 7 agonists
Abstract
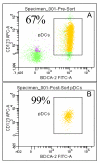
Background: Plasmacytoid Dendritic Cells (pDC) comprise approximately 0.2 to 0.8% of the blood mononuclear cells and are the primary type 1 interferon (IFN), producing cells, secreting high levels of IFN in response to viral infections. Plasmacytoid dendritic cells express predominantly TLRs 7 & 9, making them responsive to ssRNA and CpG DNA. The objective of this study was to evaluate the molecular and cellular processes altered upon stimulation of pDC with synthetic TLR 7 and TLR 7/8 agonists. To this end, we evaluated changes in global gene expression upon stimulation of 99.9% pure human pDC with the TLR7 selective agonists 3M-852A, and the TLR7/8 agonist 3M-011.
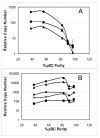
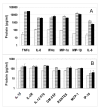
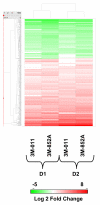
Results: Global gene expression was evaluated using the Affymetrix U133A GeneChip(R) and selected genes were confirmed using real time TaqMan(R) RTPCR. The gene expression profiles of the two agonists were similar indicating that changes in gene expression were solely due to stimulation through TLR7. Type 1 interferons were among the highest induced genes and included IFNB and multiple IFNalpha subtypes, IFNalpha2, alpha5, alpha6, alpha8, alpha1/13, alpha10, alpha14, alpha16, alpha17, alpha21. A large number of chemokines and co-stimulatory molecules as well as the chemokine receptor CCR7 were increased in expression indicating maturation and change in the migratory ability of pDC. Induction of an antiviral state was shown by the expression of several IFN-inducible genes with known anti-viral activity. Further analysis of the data using the pathway analysis tool MetaCore gave insight into molecular and cellular processes impacted. The analysis revealed transcription networks that show increased expression of signaling components in TLR7 and TLR3 pathways, and the cytosolic anti-viral pathway regulated by RIG1 and MDA5, suggestive of optimization of an antiviral state targeted towards RNA viruses. The analysis also revealed increased expression of a network of genes important for protein ISGylation as well as an anti-apoptotic and pro-survival gene expression program.
Conclusion: Thus this study demonstrates that as early as 4 hr post stimulation, synthetic TLR7 agonists induce a complex transcription network responsible for activating pDC for innate anti-viral immune responses with optimized responses towards RNA viruses, increased co-stimulatory capacity, and increased survival.
Figures




Similar articles
-
Plasmacytoid dendritic cells produce cytokines and mature in response to the TLR7 agonists, imiquimod and resiquimod.Cell Immunol. 2002 Jul-Aug;218(1-2):74-86. doi: 10.1016/s0008-8749(02)00517-8. Cell Immunol. 2002. PMID: 12470615
-
Comparison of human B cell activation by TLR7 and TLR9 agonists.BMC Immunol. 2008 Jul 24;9:39. doi: 10.1186/1471-2172-9-39. BMC Immunol. 2008. PMID: 18652679 Free PMC article.
-
Multiple sclerosis: modulation of toll-like receptor (TLR) expression by interferon-β includes upregulation of TLR7 in plasmacytoid dendritic cells.PLoS One. 2013 Aug 12;8(8):e70626. doi: 10.1371/journal.pone.0070626. eCollection 2013. PLoS One. 2013. PMID: 23950974 Free PMC article.
-
Plasmacytoid dendritic cell precursors/type I interferon-producing cells sense viral infection by Toll-like receptor (TLR) 7 and TLR9.Springer Semin Immunopathol. 2005 Jan;26(3):221-9. doi: 10.1007/s00281-004-0180-4. Epub 2004 Nov 13. Springer Semin Immunopathol. 2005. PMID: 15592841 Review.
-
IRF family proteins and type I interferon induction in dendritic cells.Cell Res. 2006 Feb;16(2):134-40. doi: 10.1038/sj.cr.7310018. Cell Res. 2006. PMID: 16474425 Review.
Cited by
-
A novel TLR7 agonist as adjuvant to stimulate high quality HBsAg-specific immune responses in an HBV mouse model.J Transl Med. 2020 Mar 4;18(1):112. doi: 10.1186/s12967-020-02275-2. J Transl Med. 2020. PMID: 32131853 Free PMC article.
-
Transcriptional profiling of intrinsic PNS factors in the postnatal mouse.Mol Cell Neurosci. 2011 Jan;46(1):32-44. doi: 10.1016/j.mcn.2010.07.015. Epub 2010 Aug 7. Mol Cell Neurosci. 2011. PMID: 20696251 Free PMC article.
-
TLR7 stimulation in human plasmacytoid dendritic cells leads to the induction of early IFN-inducible genes in the absence of type I IFN.Blood. 2009 Aug 27;114(9):1794-802. doi: 10.1182/blood-2009-04-216770. Epub 2009 Jun 24. Blood. 2009. PMID: 19553637 Free PMC article.
-
Combination therapy targeting toll like receptors 7, 8 and 9 eliminates large established tumors.J Immunother Cancer. 2014 May 13;2:12. doi: 10.1186/2051-1426-2-12. eCollection 2014. J Immunother Cancer. 2014. PMID: 24982761 Free PMC article.
-
Regulated cell death and DAMPs as biomarkers and therapeutic targets in normothermic perfusion of transplant organs. Part 1: their emergence from injuries to the donor organ.Front Transplant. 2025 Apr 24;4:1571516. doi: 10.3389/frtra.2025.1571516. eCollection 2025. Front Transplant. 2025. PMID: 40343197 Free PMC article. Review.
References
-
- Zhang Z, Wang FS. Plasmacytoid dendritic cells act as the most competent cell type in linking antiviral innate and adaptive immune responses. Cell Mol Immunol. 2005;2:411–7. - PubMed
-
- Yoneyama H, Matsuno K, Zhang Y, Nishiwaki T, Kitabatake M, Ueha S, Narumi S, Morikawa S, Ezaki T, Lu B, Gerard C, Ishikawa S, Matsushima K. Evidence for recruitment of plasmacytoid dendritic cell precursors to inflamed lymph nodes through high endothelial venules. Int Immunol. 2004;16:915–28. doi: 10.1093/intimm/dxh093. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

